Hi, I would like to trouble shoot given several issues with the tutorial SEEG epileptogenicity maps. This is my brainstorm version
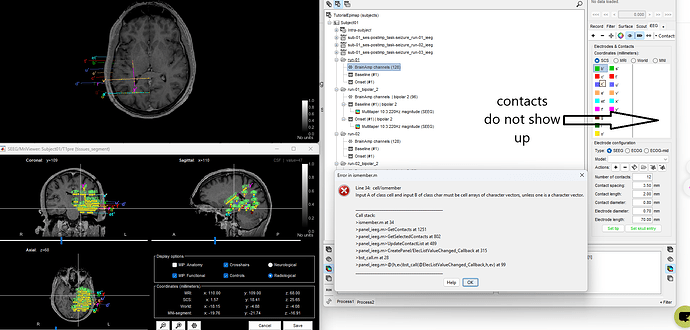
The first issue is the Add EEG positions does not allow contacts to be reviewed even though the electrodes appear in the iEEG panel.
Please see below. I presume this is because the .tsv file provides selected contacts and not contacts based on an SEEG electrode model. This is first issue will post other issues in that tutorial
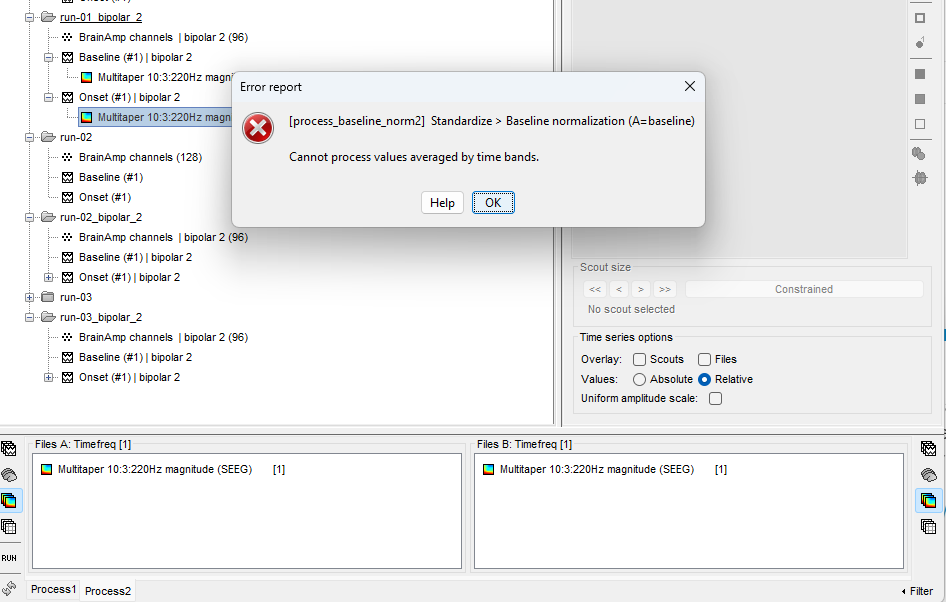
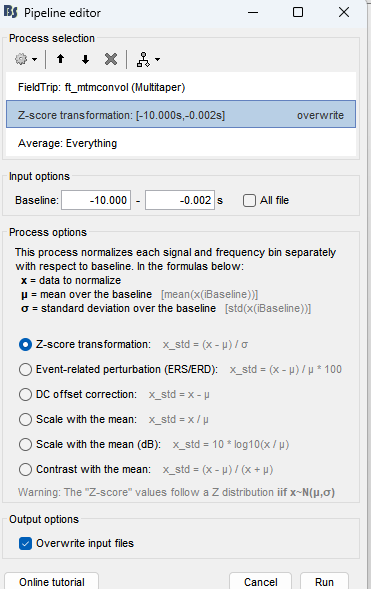
second issue i am having is baseline normalizing as per the Time-frequency analysis (separate baseline) of the tutorial
this is the error i am getting
video for more details
https://1drv.ms/v/c/cf269046e55bcbb0/EVNaQrMzuqVHvEmuijib1ewBYn9whu3ngQpUu6SHjA3Lvg?e=UECw2s
@mubafzal2 thank you for reporting.
From the screenshot it seems you are not on the latest version of Brainstorm (see image below for the current version)
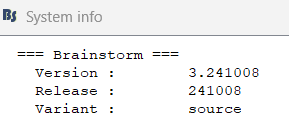
Can you update your instance and see if the errors still persist ?
Hi chinmay thanks for getting back. The contact identification works now with the new version update and brings out the contacts. however the time-frequency steps is not working. the three steps protocol in section Time-frequency analysis (pre-onset baseline) and the Time-frequency analysis (separate baseline).
@mubafzal2 this error report was it generated running the tutorial_epileptogenicity('path to tutorial_epimap_bids) command as mentioned here ? If not, can you maybe explain the steps in details.
I am unable to reproduce it at my end.
FYI @Raymundo.Cassani
Hi Chinmay,
its these steps of the tutorial
- In Process1, select all the Onset bipolar files.
- Select the process: Frequency > FieldTrip: ft_mtmconvol (Multitaper): (do not run)
- Time window: [-10, 10]s
- Sensor types: SEEG
- Taper: hanning
- Frequencies: 10:3:220
- Modulation factor: 10
- Time resolution: 1000ms
- Time step: 100ms (rounded to 99.6ms because of the sampling frequency)
- Measure: Magnitude
- Save average: Disabled
- Add the process: Standardize > Baseline normalization:
- Baseline: [-10, -1]s
- Method: Z-score transformation
- Overwrite: Enabled
- Add the process: Average > Average files:
- Group files: Everything
- Function: Arithmetic average
after importing onset and baseline and doing this
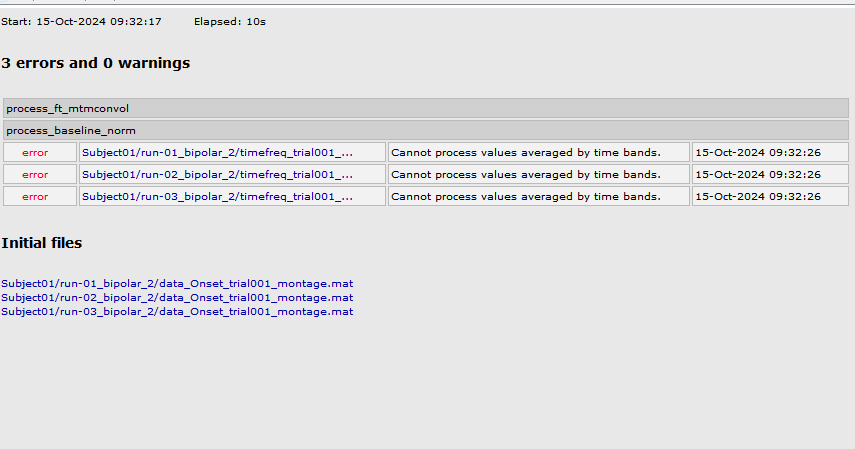
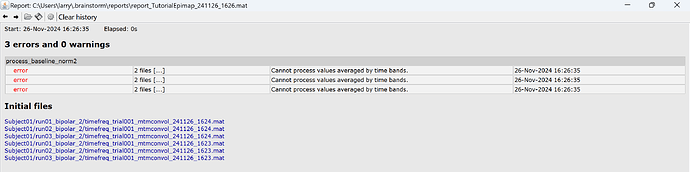
I get error this
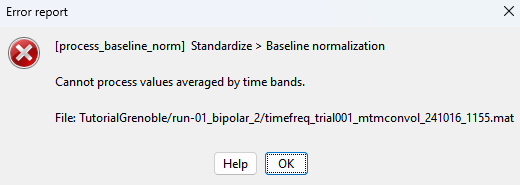
btw the epilepogenicity map section of tutorial is woking well with the change in brainstorm version from 241003 to 241008 now but the tfr decomposition step still
not working well as yet
Hi @mubafzal2, the error that you report is expected due to a change in the data for the TF map from multitatper. Please update your Brainstorm instance to have this issue fixed.
More details: since the TF maps from multitaper are not "continuous" (same time definition as the sensor time series), in June 2024 (commit: 4b0d9782), we updated the TF structure for multitapers so they are time-band defined. So it is possible to plot them along side their time series, this was an error reported in the past.
Hi, I am having the same issue with baseline normalization. The version of my brainstorm is 3.241122 (22-Nov-2024) and issue with the code of TF maps has already been fixed. The following are the details of my operations and screenshots:
- In Process1, select all the bipolar files (Onset and Baseline).
- Run the process: Frequency > FieldTrip: ft_mtmconvol (Multitaper):
- Time window: 0 - 5s All file
- Sensor types: SEEG
- Taper: hanning
- Frequencies: 10:3:220
- Modulation factor: 10
- Time resolution: 1000ms
- Time step: 99.6ms
- Measure: Magnitude
- Save average: Disabled
Run process1
In Process2,Select the process: Standardize > Baseline normalization (do not run immediately):
- Baseline:0.5-4.48s All file
- Method: Z-score transformation
Add process: Average > Average files:
- Group files: Everything
- Function: Arithmetic average
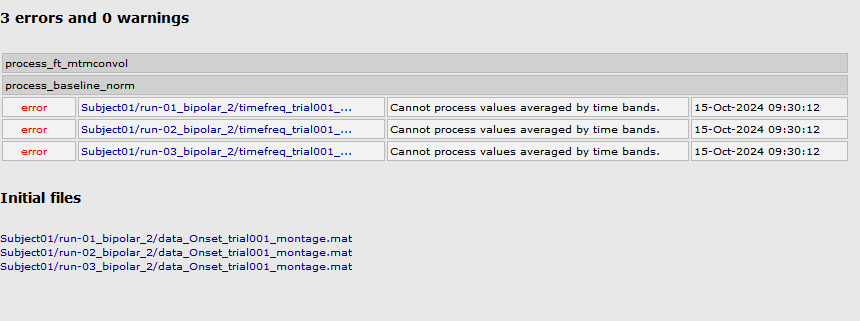
After run the pipeline I got the same error:
And I am using the data from the tutorial.
Thanks for help.
@LanLingXiaoXiaoSheng, This error was a consequence of the update on the time-definition for TF files from multitaper as described in here: SEEG epileptogenicity tutorial - #7 by Raymundo.Cassani
Now it is addressed for processes in the Process1 and Process2 tabs. Commit da16c5f
Please update your Brainstorm instance, and try once more.
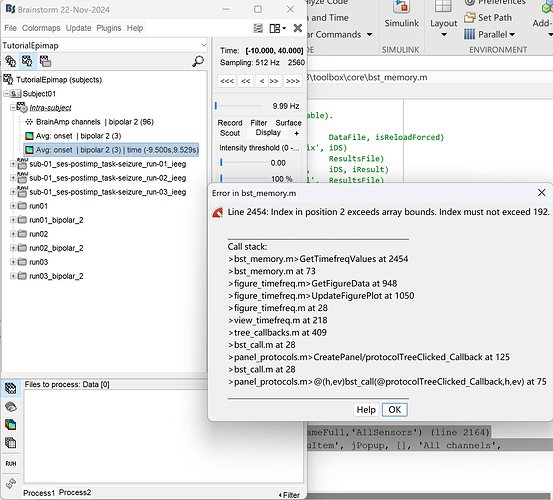
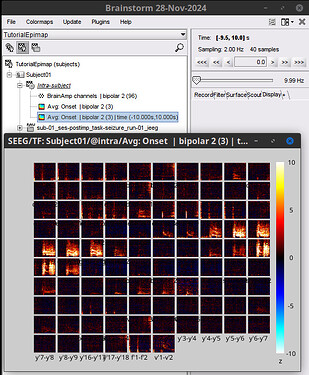
Thank you for your help. The issue with baseline normalization has been resolved, but now I have encountered a new problem. When I try to display the time-frequency plot for all channels, MATLAB shows this:
The screenshot shows you are using the 22-Nov-2024 version.
Please use 26-Nov-2024 or later, where this was fixed, as per my previous message:
You can trigger the update with the menu Update > Update Brainstorm in the main window