Then, I used the "aseg atlas" from the default anatomy and combined it with the generated cortex from CAT12.
This is definitely not a good idea.
If Brainstorm can generate "cortex_cereb" without FreeSurfer, is there a way to do it without using the default anatomy?
I added a new menu "Create surfaces", when you right-click on an atlas: Anatomy: Create surfaces from MRI volume atlas · brainstorm-tools/brainstorm3@efb5c4a · GitHub
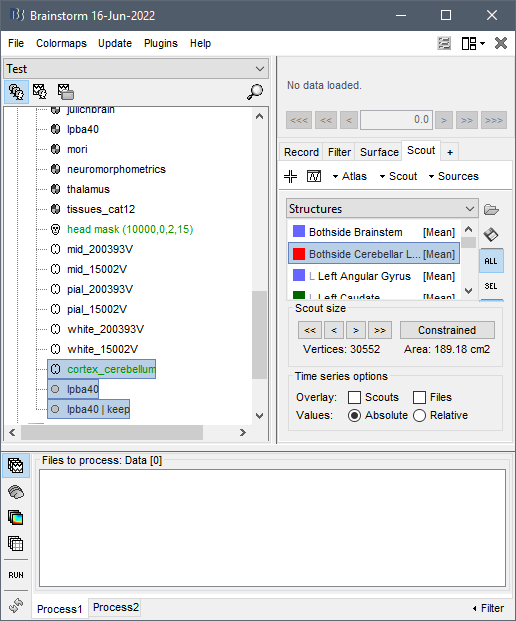
- Start by updating Brainstorm.
- After the CAT12 segmentation (generating the volume atlases), right-click on the volume atlas
lpba40> Create surfaces. - OPTIONAL: Downsample the new surface file
lpba40(it depends how many sources you want there) - Double-click on the new surface file
lpba40 - In the Scout tab, select atlas "Structures" in the drop-down menu
- In the 3D figure, click on the cerebellum, it should select the scout
Bothside Cerebellar Lobe - In the Scout panel, with this scout selected, select menu Scout > Edit surface > Keep only selected scout
- In the database explorer, select simultaneously the last surface created and the cortex you want to use for source estimation, right-click > Merge surfaces
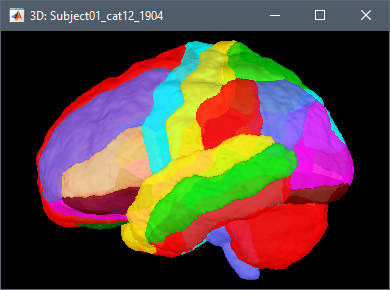
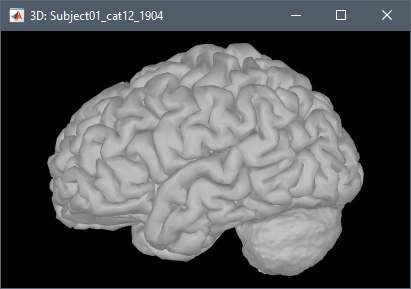
Note that this is not a realistic surface. It can be interesting in a surface-based source model to add dipoles in the cerebellum to be able to represent sources coming from the cerebellum and/or from the neck muscles (e.g. https://neuroimage.usc.edu/brainstorm/Tutorials/RestingOmega#Power_maps) in order to reconstruct cleaner activity on the cerebrum cortex. However, this surface is not adapted to represent correctly the activity of the cerebellum, as it does not include dipoles inside the cerebellum.
If you are interested specifically in the activity of the cerebellum, you might prefer using a volume source model.
https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource