Hello!
I want to evaluate crown device for source modelling. It contains 8 EEG sensors CP3/4, F5/6, C3/4, PO3/4. All I am trying to do is to simulate EEG signals based on the sensors locations of crown and then try to see the results of source modelling comparing with 19 eeg electrodes or more how do they differ with the other density of eeg and in which regions it affects the results the most. If there is any suggestion that could be helpful I will appreciate it.
I know how to modify montage file. Also I can do source modelling. My question is if it is better volume or surface head models? How can I compare the results of one density to another?
Thanks for all the help in advance!
Where are the EEG signals coming from? A full cap recording? Are they simulated?
-
Regardless the origin of the simulated EEG signals, you should create a new folder for the highest resolution cap. This folder will contain the channel file and recordings file. Verify that all the EEG channels have EEG as Type, right-click on the channel file > Edit channel file.
-
To create the lower-resolution folders, duplicate the high-resolution folder (right-click on folder > File > Duplicate folder). Change the Type of unwanted (extra) EEG electrodes, from EEG to MISC. Repeat these step for the different low-resolution folders.
-
At this step, you can double-check the EEG caps for each of the different resolution folders. For each folder plot the sensors over the head, right-click the channel file > Display sensors > EEG (head)
-
Compute the head model (forward model), and the noise covariance matrix for each folder.
-
Perform source estimation for each of the folders. Since all folders belong to the same Subject, the same anatomy is used, thus the sources can be compared as they are in the same space.
Regarding surface and volume source space. Which one is better depends on your research question.
Check more details in here:
https://neuroimage.usc.edu/brainstorm/Tutorials/HeadModel#Source_models
1 Like
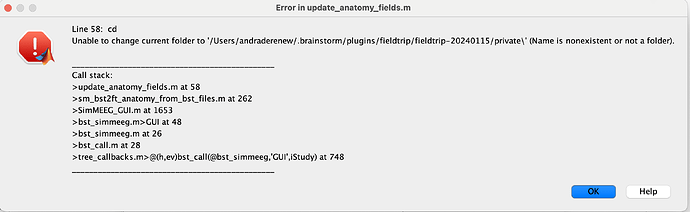
I have tried but I'm stuck in the step of obtainaing a valid source estimation. I obtain an empty file. I have no covariance matrix. May be is that. I tried SimMEEG but I think I obtain an error because there is a problem with name of folder.
The issue is the OS, there is a hardcoded path separator (for Windows) in the update_anatomy_fields.m in SimMEEG. Open such file and remove \ after 'private' in the line 58.
1 Like