Dear Brainstorm team,
I am trying to perform a functional connectivity analysis of resting state EEG data in source space (ROI level). There are currently two datasets planned to be used: dataset 1 (with individual T1 MRI), and dataset 2 (with individual defaced T1 MRI).
Preprocessing pipeline of dataset 1 (brief):
Filtering: 1-45 Hz -> downsampling to 250 Hz -> removal of bad segments and channels -> interpolation of bad channel signals -> average reference -> ICA -> removal of artificial components
Preprocessing pipeline of dataset 2 (brief):
downsampling to 250 Hz -> filtering: 1-45 Hz -> removal of bad segments and channels -> ICA -> removal of artificial components.
The duration of each recording is about 3 minutes for dataset 1 and 8 minutes for dataset 2.
I am trying to use Brainstorm for resting-state EEG source localization and encountering several problems as follows:
-
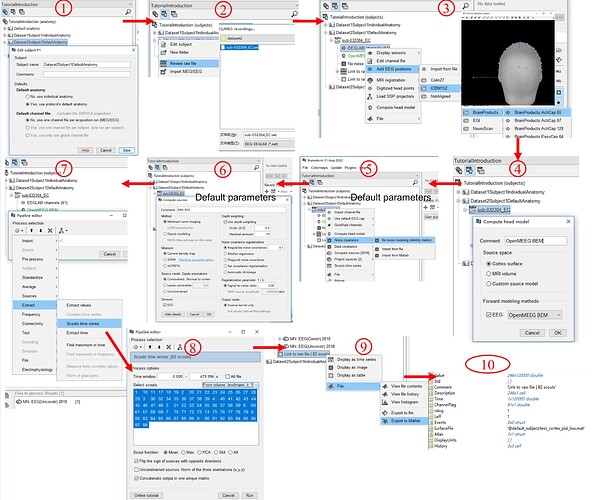
I first use the default anatomy and tried the whole process, and I wonder if the whole process is reasonable (as shown in the figure below).
-
Dataset 1 does not record the true electrode positions. Dataset 2 (i.e., the public dataset) provides digitized EEG channel locations ("Polhemus PATRIOT Motion Tracking System (Polhemus, Colchester, VT, USA) localizer together with the Brainstorm toolbox was used to digitize the exact location of each electrode on a participant's head relative to three fiducial points. "). After the import of individual T1, the fiducial points (AC, PC, IH) were determined automatically, but the precision did not seem to be sufficient (as shown in the figure below), so I wonder if I need to set them manually again. For dataset1, is manual alignment the only available method if using individual T1? when importing digitized EEG channel locations for Dataset2Subject1, all electrodes are above the scalp and not attached to the scalp. In addition, only the defaced T1 is available, it seems to adversely affect the setting of fiducial points, co-registration of electrodes with MRI, MRI segmentation, and source localization.
Figure 2, please click the following link (since new users can only put one embedded media item in a post):
清华大学云盘 -
After MNI parcellation is downloaded from Brianstorm (i.e., Brodmann atlas), is the atlas automatically aligned to individual space by Brainstorm?
-
As mentioned in the preprocessing pipeline, the bad channel signals are interpolated (Dataset1), the interpolation does not add new information. But if this will have a bad effect on source localization or not. A related issue is on dataset2: ICA removed a large number of independent components, for example, there are 60 in the original and 20 in the final remaining, that is, the rank of this recording is only 20 after preprocessing, I wonder if source localization results of such data are reliable?
-
Dataset 1 is re-referenced to average, and the reference electrode of Dataset 2 is FCz. Is there a recommended reference method? In addition, sometimes, the EEG data are spatially filtered (surface Laplacian) to reduce the effect of volume conduction effect in calculating the channel-level connectivity, I wonder if the spatially filtered data can be used for source localization.
-
In source files, there is a variable ImagingKernel, can source time series be directly obtained by ImagingKernel (dimension: Nsources x Nchannels) × the recording (dimension: Nchannels x Timepoints)? Will variable Whitener be involved in this?
-
One more important question is about the sign of the sources. For ROI-based analysis, there seem to be two often used methods: one is to flip dipoles with the opposite signs from the orientation and then average the signals within the ROI; the other is to find the first principal component of the source signals within the ROI. Does the Brainstorm team have any suggestions and recommendations for this issue? In addition, due to the uncertainty of the sign of the source signal, although the signal at the ROI level can be obtained by the above two methods, it seems still uncertain whether the true signs of the signal are the same or opposite between different ROIs. So is this still possible to calculate the phase-synchronization-based functional connectivity between ROIs (e.g., phase-locked value or phase lag index)? A reference mentions that most phase information can be preserved after taking the absolute value of the source signals (see the figure below), I am not able to make a judgment on this. Can the source signals (dimension: 246×120000) obtained in steps 8-10 of the pipeline be used directly to calculate the functional connectivity (e.g., PLI)?
Figure 3, please click the link in question 2 (since new users can only put one embedded media item in a post). -
The number of channels for both EEG datasets is about 60. Does the Brainstorm team have any recommendation on whether to use BEM or FEM header model, to construct constrained sources or unconstrained sources? In addition, based on the responses from the Brianstorm team to others, it seems that the current density map and dSPM are more recommended than sLORETA for source localization.
-
If all subjects are using the default anatomy, then the source signals are in the standard MNI space, and no registration between subjects is involved. However, if individual T1 is used, the current process seems to end up with the alignment of the atlas (e.g., Brodmann) to individual T1 (individual space), and then the signal of each ROI is extracted. If group analysis will be performed, it seems that the step of mapping the source signals of each subject to the standard space is missing. Another method I thought of is to first align the T1 of all subjects to the standard space, then perform EEG source localization. However, I wonder if source localization using individual T1-> alignment to standard space and alignment individual T1 to standard space -> source localization are equivalent?
-
Can the source localization process as shown in Figure 1 be fully implemented by MATLAB codes?
Example data for both datasets can be downloaded directly from the following link:
https://cloud.tsinghua.edu.cn/f/433d489f944c43ecacd2/?dl=1
Thank you very much for your help!
Best regards,
Milton