Hi Brainstorm,
I want to do a group analysis on source models, but I worry I am doing something wrong. I have some questions on averaging source models and scouts time series.
Some background: I am doing an analysis on somatosensory evoked potentials (SEPs) in MEG data. I used the default anatomy for my subjects and warped the anatomy for each of them. For this question, I am looking at surface source models.
First, I averaged my epochs based on the SEP triggers for each separate session (sensor average). Then, as I am using the default anatomy, I used Unconstrained source estimation for all sensor averages (Settings: Compute sources [2018]. Minimum norm imaging, Current density map, Unconstrained, MEG sensors.). (As I am working with MEG data, I used overlapping spheres (surface) as head models.)
According to the Workflows tutorial (https://neuroimage.usc.edu/brainstorm/Tutorials/Workflows#Unconstrained_cortical_sources) for group analysis, the next step for a group analysis is to normalize (z-score) the current density source maps for all subjects w.r.t. baseline. I did this with the Process1 box (Process sources > Standardize > Baseline normalization. Baseline 200 to 5 ms before SEP trigger. Z-score). Next, I flattened the cortical maps (Process sources > Sources > Unconstrained to flat map. Norm). After this, I selected the normalized and flattened source models I wanted to average and dragged them into Process1. I used Process sources > Average > Average files > Everything. I used 'Everything' as I only dragged the source maps I wanted to average into the Process1 box.
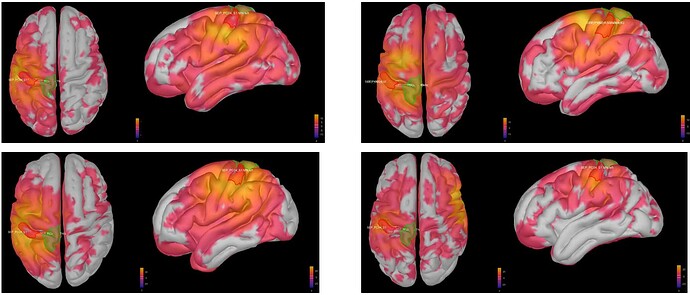
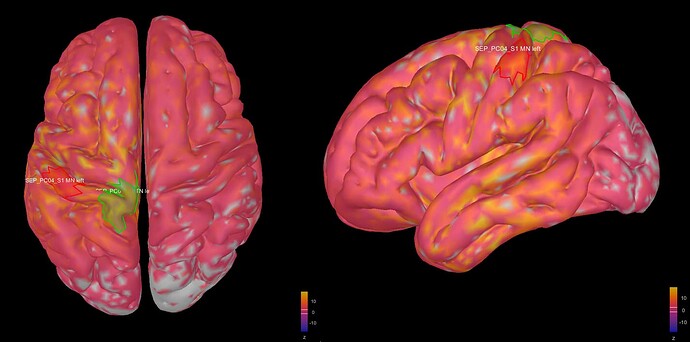
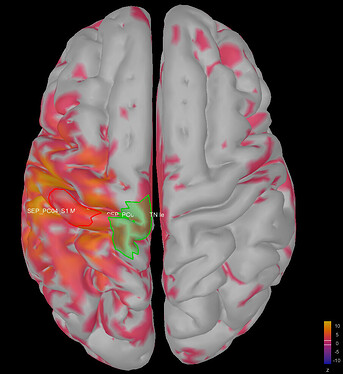
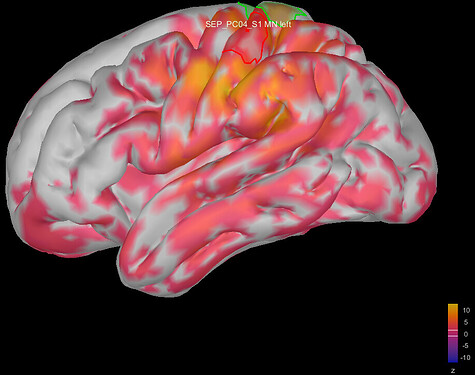
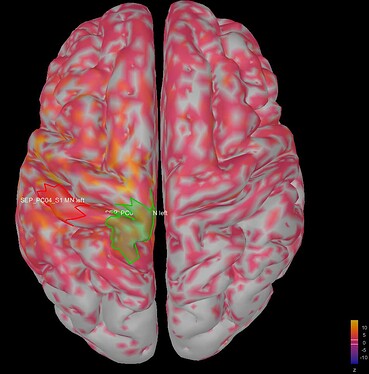
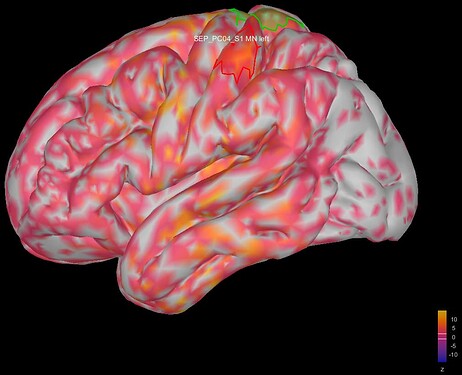
This is where I was surprised: the average source model looks very messy and speckled, whereas the individual source models of my subjects looked very clean. Next to this, the focus of activity that I am expecting in the primary somatosensory cortex (like in my individual subject's source models) has spread/leaked onto almost the entire left hemisphere. I included some images below for clarity.
Is this something you see more often, and is it just part of averaging source models, or did I do something wrong here? Can it be caused by the warning I receive when averaging the source models ("The channels files from the different studies do not have the same number of channels. Cannot create a common channel file")? Should I fix this by creating a uniform channel file?
My last question is on averaging scouts time series. In case this is just the way my averaged source models will look, it is not useful for visualization of the averages and it might be better to just average my subject's scouts time series directly. So: is averaging source models and extracting the scouts time series from the average source model equal to extracting the scouts time series from subject's source models and average these scouts time series myself?
Please let me know if I need to clarify my question(s) any further. And of course, thanks in advance for helping me!
Best,
Janne
Image 1: the individual source maps of my subjects (n=4), all normalized and flattened, at t=27 ms
Image 2: my average source map of these subjects, at t=27 ms as well. They are not smoothed, smoothing did not make things better.