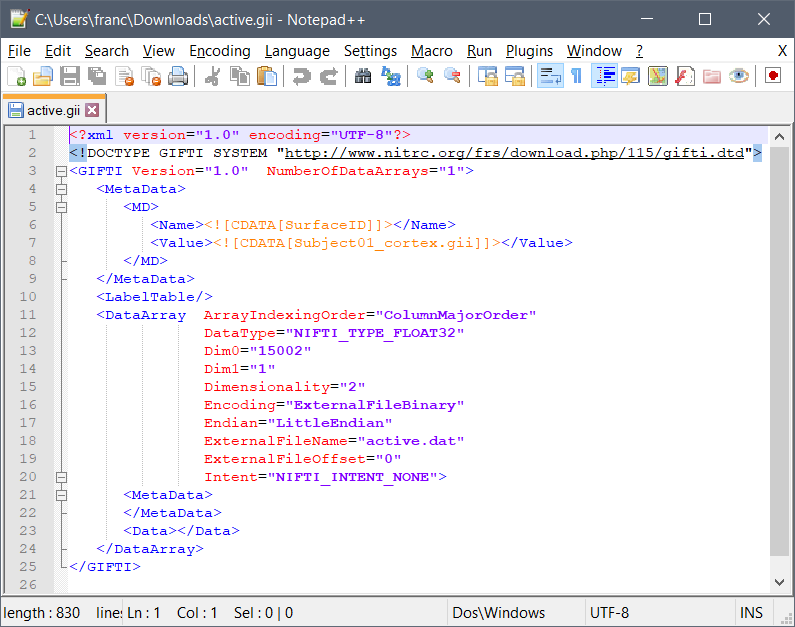
Hello Brainstorm Forum.
@ZaidaEMtzMo , other lab members and I have been trying to compare our novel MEG tonotopy mapping technique with an established fMRI technique. When we compared the results, we noticed that the anatomy differed between SPM/AFNI/SUMA and Brainstorm. Through emails, @Francois suggested to modify some of the Brainstorm projection scripts, but we didn't feel confident enough to open Brainstorm's hood right now.
We decided to instead try to project our MEG results through SPM/AFNI/SUMA, but we faced multiple issues trying that. Here's a list of the methods attempted but that didn't yield proper results. Detailed issue description for each can be found bellow.
- Using the 'Export to SPM8/SPM12 (volume)' process and opening in AFNI/SUMA
- Using the 'Export to SPM8/SPM12 (volume)' process and opening in SPM12
- Using the 'Export to SPM12 (surface)' process and opening in AFNI/SUMA
- Using the 'Export to SPM12 (surface)' process and opening in SPM12
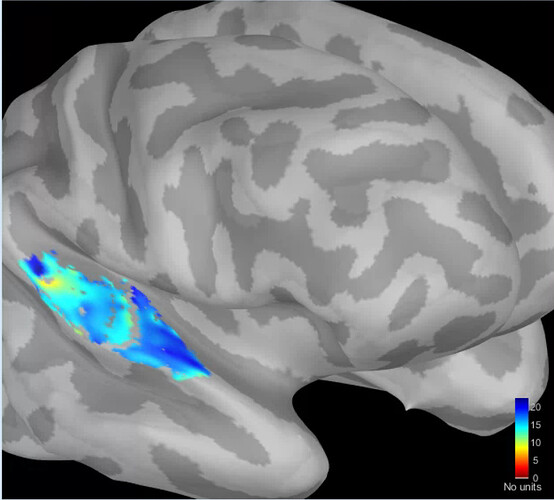
The original Brainstorm surface looked like this:
At first, we kept the default settings. It lead to a very pixelated situation given that there was a spatial downsampling option.
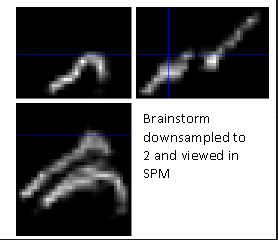
Following this, we removed it by setting the downsampling to 1.
Here are the details on each individual attempt:
-
Using the 'Export to SPM8/SPM12 (volume)' process and opening in AFNI/SUMA
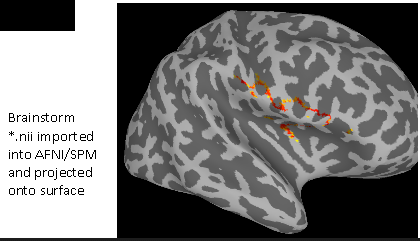
-
Using the 'Export to SPM8/SPM12 (volume)' process and opening in SPM12

As we can see, either software display a strange, stringy thing instead of the expected islands. From these two results being similar, we thought that perhaps the 'Export to SPM8/SPM12 (volume)' process didn't work as we expected and tried the other process. In the end, we want to compare surfaces anyway.
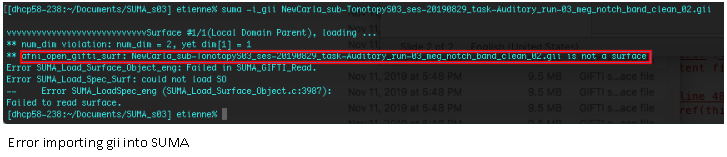
- Using the 'Export to SPM12 (surface)' process and opening in AFNI/SUMA
The issue seems to be coming from the .gii file type. AFNI/SUMA can'y open it.
Perhaps AFNI/SUMA does not recognize that file type, so we moved on to SPM12.
- Using the 'Export to SPM12 (surface)' process and opening in SPM12
Similarly, SPM12 was not able to open the .gii file.

We were wondering if we are using these processes wrongly. Maybe someone from the Brainstorm community could give us a hand in solving this issue.
Thank you
1 Like
Converting between coordinates systems is always a nightmare.
Unfortunately we don't have any experience with AFNI and very limited knowledge of SPM, we might not be able to help you much in your project. But I can at least try to help you in evaluating if there is any issue in what Brainstorm exports.
-
Volume approach: make sure that what you get out of Brainstorm makes sense:
Overlay the .nii obtained with Brainstorm with an MNI template in MRIcron (safer option to test transformation matrices and coordinate systems than Brainstorm or SPM).
-
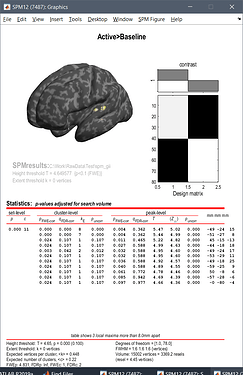
Surface version: Try to replicate what is done in the SPM12 export tutorial (using the dataset from the introduction tutorials: deviant condition, baseline=[-20,-15]ms, active=[85,95]ms)
https://neuroimage.usc.edu/brainstorm/ExportSpm12
(update SPM12 before testing)
I've just tested it and it works as expected, leading me to think there is no problem with the .gii files generated with Brainstorm:
Perhaps AFNI/SUMA does not recognize that file type, so we moved on to SPM12.
The export to a SPM12 surface creates two .gii files: the cortex surface and a texture on this surface with the source values (which is indeed NOT a surface .gii file, it does not contain vertices or faces). This is explained in the tutorial. Make sure you didn't get mixed up.
Hello Francois,
Thank you for your sustained help. We used your advice to progress on both methods but still hit roadblocks.
Volume approach:
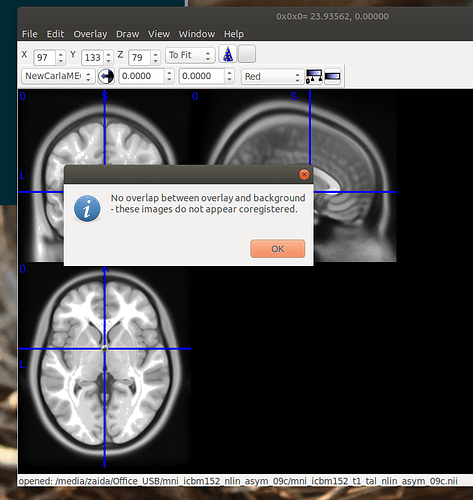
We managed to open the strange output in MRIcron. When we tried to overlay the MNI template, we had an error saying that the files are not co-registered.
Surface approach: We did get confused with the two .gii, as you predicted. Once that was fixed, we tried to follow the ExportSpm12 instructions but we are under the impression that it doesn't apply exactly to our dataset. The instructions seem to be more focussed on stats analysis, whereas we merely want to see the surface. We don't have a "baseline" either in this condition. Given that, we were not able to reach the surface visualization.
We also tried to open the correct .gii in SUMA. It worked, but we didn't have the option to inflate the brain to match the previous figure. We will post on their forum to see if something can be done from that end.
Thanks again
Why strange? You have estimated the surface on a small ribbon (the cortex envelope), when reinterpolating the values on the volume, you don't get any values outside of that ribbon.
If you want full volume maps, use a volume source model:
https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource
When we tried to overlay the MNI template, we had an error saying that the files are not co-registered.
I've just checked again, and it seems to be working correctly.
Try to reproduce the following:
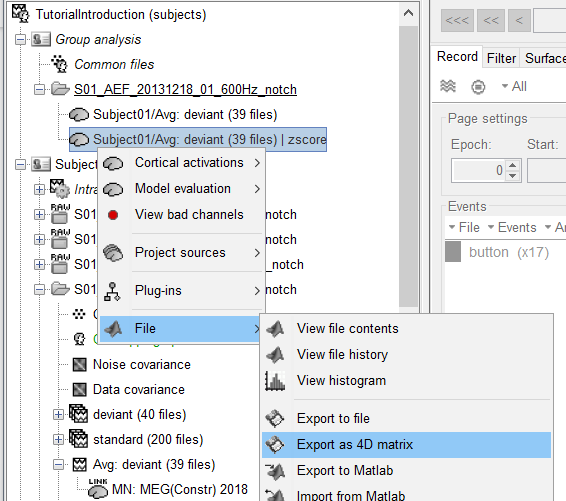
- In the protocol you created with the introduction tutorials: right-click on the deviant average MNE sources > Project > Default anatomy > Cortex 15002V
- Run a Z-score normalization on projected source maps (baseline = [-100,0]ms)
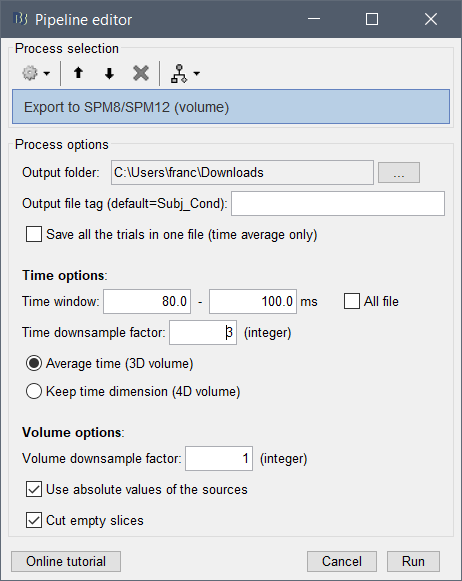
- Right-click on the normalized projected sources > Export as 4D matrix (options displayed below)
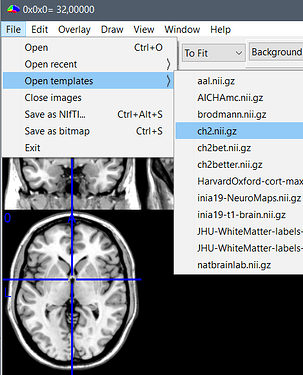
- In MRIcron: Menu File > Open template > ch2
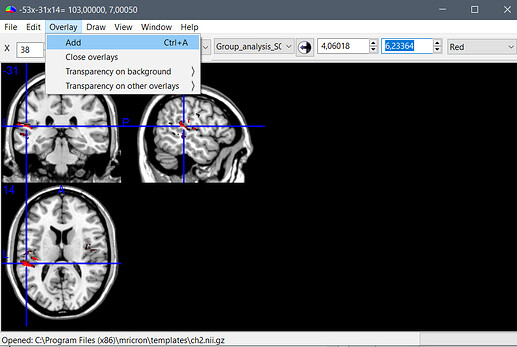
- Then menu Overlay > Add > Select the file that was exported by Brainstorm, and adjust the colormap settings (see the result below)
Surface approach:
I'm sorry, I'm not sure what to tell you beyond that. The export to .gii seems to be working and the output files can be read and displayed in SPM correctly. Unless you find any obvious error in Brainstorm, I don't know what else I can do for you.
Hello again Francois,
Thanks for your explanation, we understand why the volume approach is getting us only a ribbon of data. We will try to avoid it, else we will have to play with co-registrations and we don't have much expertise concerning that either.
Concerning the surface approach, we reached out to the AFNI/SUMA people and they are willing to help us. We shared the output of ExportSpm12 and would like to give them a bit of detail about it, but I need a little bit of help.
On the tutorial page we find these description for the three output files:
- Subject01_cortex.gii: Contains the definition of the cortex surface (faces and vertices)
- subject01_left_active_*.gii: 101 files, definition of the trials for the active condition
- subject01_left_active_*.dat: 101 files, actual source maps for the active condition
Other than the first one, we aren't too sure what these mean as we can't open or read them to investigate more. We'd be very grateful if you could teach us or point us a to recourse with more details.
Once again, thank you for your help,
Thanks for your explanation, we understand why the volume approach is getting us only a ribbon of data. We will try to avoid it, else we will have to play with co-registrations and we don't have much expertise concerning that either.
You would have the same problems of coregistration whether you use a surface or volume source estimation... Ribbon or no ribbon should change your processing pipeline, it's just a way of representing the source activity.
Other than the first one, we aren't too sure what these mean as we can't open or read them to investigate more. We'd be very grateful if you could teach us or point us a to recourse with more details.
These are textures in .gii files, one value per vertex in the corresponding cortex mesh. You can open the .gii file with any text editor.
The full specification: NITRC: GIFTI: Tool/Resource Info