Hi BST experts,
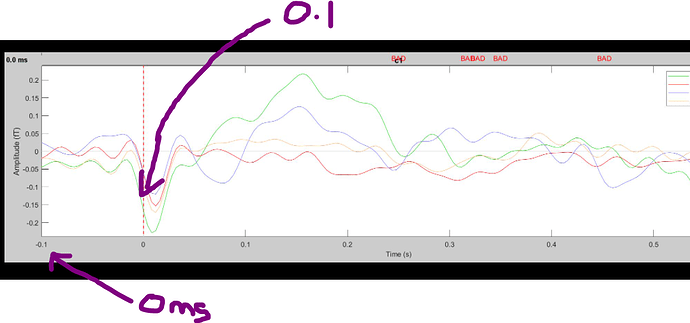
When I display time series graphs, the zero point looks to be positioned around where I expect to see activity occurring at 100ms - i.e., the timing looks to be lagging by around 100ms. Passive auditory response data were obtained with whole cortex 160 channel MEG (coaxial, first order gradiometers - Model PQ1160R-N2, KIT), and epoched at -100 to 600ms around events. The plot below is for grand averages (2 groups x 2 conditions) with arrows indicating the timing issue. The delay occurs with individual plots as well. I'm not sure if this is an averaging error? Epoching error? Or something else. There was no systematic stimulus delay and I'm not sure of the precise timing difference, so manually adding a time offset is not a possibility. I'm hoping someone can help resolve this issue so I can make sure the final averages used in analysis are accurate.
Many thanks in advance.
The time zero is set at the exact latency of the event markers you epoched you data from.
https://neuroimage.usc.edu/brainstorm/Tutorials/Epoching
You need to check the latency of these events in the continuous recordings, before epoching.
And you should definitely measure precisely the stimulus delays between the trigger in your file and the actual presentation of the stimulus to the subject. Without an exact measure of the presentation delays, you can't expect to have accurate latencies in your ERPs.
https://neuroimage.usc.edu/brainstorm/Tutorials/StimDelays
Thanks Francois,
We have checked the continuous recordings (raw .con converted to .csv for Brainstorm), and the timing is correct for all participants, with the same correct latency between each stimulus presentation as expected.
We have also measured and compensated for stimulus delays in the trigger and actual presentation on the MEG (Kit-Kanazawa system at Macquarie in Sydney) after data recording, before converting it for Brainstorm analysis.
It looks like there is a systematic 100ms delay in all of our data post-epoching at Averaging -where we expect 0 - 100ms is where brainstorm labels -0.1ms to 0ms.
We've also plugged the data into BESA and SPM and the Averages are correct (e.g. 100ms peak is as expected at 100ms). So we wonder if there is something different in the Averaging or post-ICA that you can help us to pinpoint?
Would really appreciate any insight on this, or where we can interrogate which steps that may have been missed or incorrectly entered.
Thanks again in advance.
I guess your problem is mostly coming from your understanding of the inputs of the epoching options (menu Import MEG/EEG). If your marker you use for epoching is the beginning of the epoch and not the stim (i.e. 100ms before the stim), just changing the "epoch time" from [-100ms,600ms] to [0ms,700ms] would fix your timing issue.
https://neuroimage.usc.edu/brainstorm/Tutorials/Epoching
If you want to fix the timing for existing files, use process "Pre-process > Add time offset".