Hello Experts!
I had segmented MRI using CAT12. I have used volume conductor head model template grid which has 1645 grids. As described in https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource#Volume_atlases. I tried importing AAL3/ASEG from default subject anatomy.
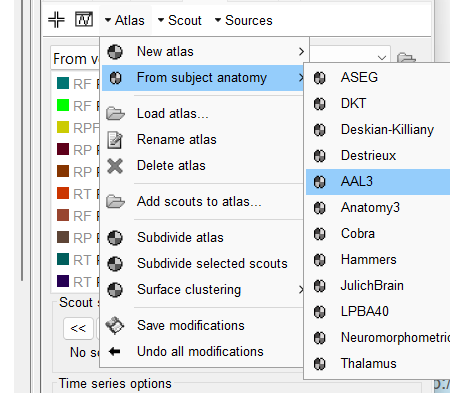
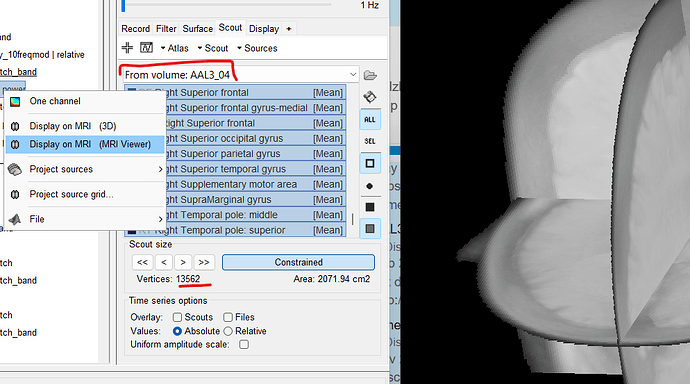
I see the identifier as "From volume:AAL3_04" - I understand it is volume based scouts.
However, when I select all the scouts, the sum of vertices turns out to be 13562. I am bit confused, if its volume based scouts - shouldn't it be using the 1645 volume grids instead of 15000 cortical surface vertices ? Can you help me understand this gap ?
Thanks!
It is because you created a surfaceAtlas from a volume anatomy parcellation (aka atlas). Because the surface source file was open. This can be deducted with these details:
- The name says: From volume: "atlasName". When a volume atlas is created from a volume parcellation, the name is always Volume "nVertexVolumeGrid": AtlasName. As indicated in the provided link
- As you can see the Scout size has a Area cm2 value, this indicates that those are surface scouts, not volume. With volume scouts, it appears Volume cm3
- Because the scouts are surface scouts, they are not displayed on the 3D MRI slices
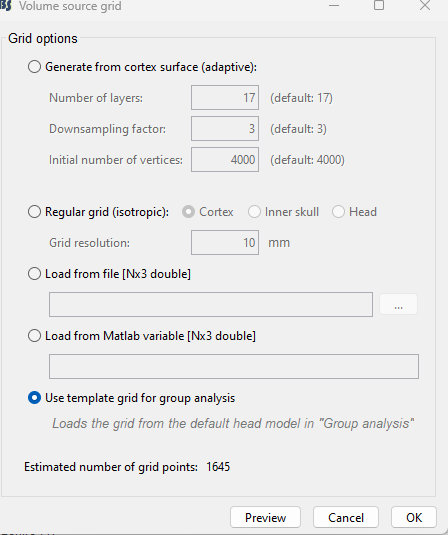
Close all the figures. Open the volume sources, then proceed to add the a volume atlas, from the anatomy.
About the total number of vertices when selecting all the Scouts, they can be less than the vertices in the surface (or volume) as there are vertices that do not correspond to any Scout
Hi Raymundo,
Got it ! Thank you for your help! I observed that for a subject the number of volume scout regions are 'p' and grip points are 'x'. When I right click on the volume sources and project it to default subject, the number of volume scout regions are 'q' and grip points are 'y'. Where p>q and x>y.
For eg: These regions {'Lobule VI of vermis';'Right Intralaminar';'Right Substantia nigra-pars reticulata'} were only in 'p' with only 1 grid point each. Is it because - they are so small that they had a corresponding grip point in the subject whereas in default subject-these scout regions had none ?
Please provide more context so we can assist you better. Add relevant screenshots
I'm not sure volume sources are being used. Because if using volume source, the option Project source grid should not change the number of vertices, because either:
- A template grid was used, so the there is the same number of vertices for the Group analysis and the Subject. Or,
- A template grid was not used, so the Subject grid is being used but in MNI coordinates to present the results on the Default anatomy
Hi Raymundo,
I have calculated the inverse model from Grid_26/3
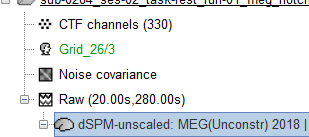
which was generated as :
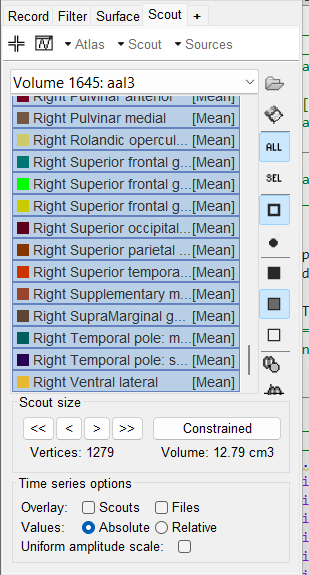
I opened the inverse model and selected Atlas -> From anatomy -> AAL and see that there are total 1279 vertices in 117 scout regions.
I right click on the inverse model and select project source grid and when I open the following file
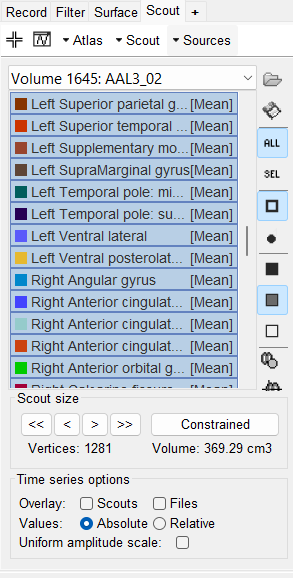
I see there are 1281 vertices in 115 scout regions.
Got it, I misunderstood the issue due to the redaction of the question.
-
The number of vertices is the same in both headmodels as they use the same grid.
-
The number of Scouts is different as they are different anatomies: There are two different anatomies: the Default anatomy and for Subject anatomy, this is to say two different MRI volumes. Thus, when those MRI are segments, their AAL3 anatomical atlas () is different for each one as, the atlas is a collection of labels for the MRI voxels, which are not the same. When the (scout) Atlas is generated (Scout tab, Atlas > From subject anatomy), each vertex in the volume is assigned with a label depending on the anatomical label of the voxel closest vertex. In your case, this process is done twice, one with the AAL3 anatomical atlas from the default anatomy and one with the ALL3 anatomical atlas from the Subject anatomy. This is the reason the results a slightly different.
I see! Thank you for the explanation!