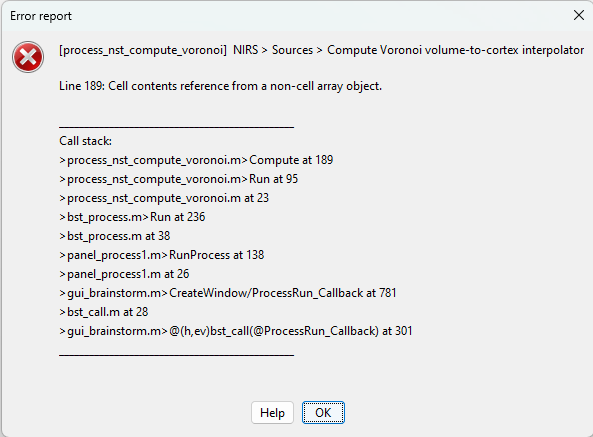
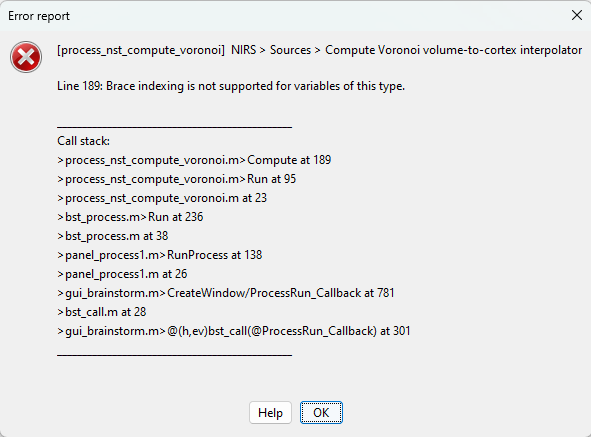
So far i was not able to reproduce.
Could you also try to execute the following script (begening of nirstorm/scripts/tutorial_nirstorn_2024.m at master · Nirstorm/nirstorm · GitHub):
(you will need to replace tutorial_dir with the path to the dataset)
tutorial_dir = "";
SubjectName = 'sub-01';
FidFile = fullfile(tutorial_dir, SubjectName, 'anat','T1w.json');
FSPath = fullfile(tutorial_dir, 'derivatives', 'FreeSurfer/',SubjectName);
TissueSegmentationFile = fullfile(tutorial_dir, 'derivatives', 'segmentation',SubjectName,'segmentation_5tissues.nii');
scoutPath = fullfile(tutorial_dir, 'derivatives', 'segmentation',SubjectName,'scout_hand.annot');
% Start a new report
bst_report('Start');
%% Part 1. Importing subject anatomical data
json = bst_jsondecode(FidFile);
sFid = process_import_bids('GetFiducials',json, 'voxel');
bst_process('CallProcess', 'process_import_anatomy', [], [], ...
'subjectname', SubjectName, ...
'mrifile', {FSPath, 'FreeSurfer+Thick'}, ...
'nvertices', 15000, ...
'nas', sFid.NAS, ...
'lpa', sFid.LPA, ...
'rpa', sFid.RPA, ...
'ac', sFid.AC, ...
'pc', sFid.PC, ...
'ih', sFid.IH);
[sSubject, iSubject] = bst_get('Subject', SubjectName);
% Import tissue segmentation
import_mri(iSubject, TissueSegmentationFile, '', 0, 0, 'segmentation_5tissues');
% Remesh skin to 10 000 vertices, iso-mesh
tess_remesh(sSubject.Surface(sSubject.iScalp).FileName ,10000 )
% Choose mid-surface as default cortical surface
iCortex = find(contains({sSubject.Surface.FileName},'tess_cortex_mid_low.mat'));
db_surface_default(iSubject, 'Cortex',iCortex);
panel_protocols('RepaintTree');
% Import hand-know region
import_label(sSubject.Surface(iCortex).FileName, scoutPath,0);
% Compute voronoi-interpolation
bst_process('CallProcess', 'process_nst_compute_voronoi', [], [], ...
'subjectname', SubjectName, ...
'do_grey_mask', 1);
ReportFile = bst_report('Save', []);
bst_report('Open', ReportFile);