|
Size: 2893
Comment:
|
Size: 2051
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| '''BrainSuite homepage has moved: '''[[http://www.loni.ucla.edu/Software/BrainSuite|click here]] <<BR>> '''BrainSuite forum: '''[[http://forum.loni.ucla.edu/forumdisplay.php?f=9|click here]]<<BR>> '''BrainSuite documentation: '''[[http://www.loni.ucla.edu/~shattuck/brainsuite/|click here]] | = BrainSuite = {{attachment:BrainSuite13_650px.png}} |
| Line 3: | Line 4: |
| = BrainSuite = {{http://brainsuite.usc.edu/images/brainsplash_sm.jpg}} |
[[http://brainsuite.org/|BrainSuite]] is a collection of image analysis tools designed to process magnetic resonance images (MRI) of the human head. !BrainSuite13 provides an automatic sequence to extract cortical surface mesh models from the MRI, tools to register these to a labeled atlas to define anatomical regions of interest, and tools for processing diffusion imaging data. !BrainSuite13 also contains visualization tools for exploring these data, and can produce interactive maps of regional connectivity. |
| Line 6: | Line 6: |
| BrainSuite is a magnetic resonance (MR) image analysis tool designed for identifying tissue types and surfaces in MR images of the human brain. The major functionality of these tools is to extract and parameterize the inner and outer surfaces of the cerebral cortex and to segment and label gray and white matter structures. BrainSuite also provides several tools for [[http://brainsuite.org/visualization|visualizing]] and [[http://brainsuite.org/interaction|interacting]] with the data. | BrainSuite is collaboratively developed between David Shattuck at the [[http://bmap.ucla.edu|Ahmanson-Lovelace Brain Mapping Center]] at UCLA and Richard Leahy in the [[http://neuroimage.usc.edu|Biomedical Imaging Group]], in collaboration with the [[http://www.loni.usc.edu/|Laboratory of Neuro Imaging]], at USC. Other contributors to the development of the methods embodied in this software include: Anand A. Joshi, Chitresh Bhushan, Justin P. Haldar, Belma Dogdas, Stephanie Sandor-Leahy, Bijan Timsari, and Paul M. Thompson. |
| Line 8: | Line 8: |
| BrainSuite is collaboratively developed between David Shattuck at the Laboratory of Neuro Imaging (LONI), UCLA and Richard Leahy in the Biomedical Imaging Group, USC. Other contributors to the development of the methods embodied in this software include: Anand A. Joshi, Belma Dogdas, Stephanie Sandor-Leahy, Bijan Timsari, and Paul M. Thompson. | !BrainSuite features include: |
| Line 10: | Line 10: |
| The currently released version of the software (BrainSuite11a) can be dowloaded here. This release includes both the graphical user interface version, available for Windows platforms, and command line versions of the the routines used for cortical surface extraction (available for Mac OSX, and Linux). The GUI can also be run on Linux and Mac OSX using [[http://www.winehq.org/|WINE]], [[http://www.kronenberg.org/wine/|Darwine (Mac OSX)]], or [[http://www.codeweavers.com/products/|Crossover (commercial)]], or using virtual machine software such as [[http://www.virtualbox.org/|Virtual Box]] or [[http://www.vmware.com/products/fusion/overview.html|VMWare Fusion]] | * Automated cortical surface extraction from T1-weighted MRI. Tools include brain surface extraction, bias field correction, voxel classification, cerebrum labeling, and surface generation * Automated surface/volume registration software * Tools for correction of B0 effects in diffusion data and coregistration to structural MRI * Tools for processing of diffusion data including ODF and tensor fitting and tractography * Sophisticated tools for visualizing and exploring MRI data, diffusion data, tractography, and connectivity |
| Line 12: | Line 16: |
| Current BrainSuite features include: | The currently released version of the software (BrainSuite) includes graphical user interface versions for Mac OSX and Windows and command line versions for Mac OSX, Windows and Linux. More details can be found on following links: |
| Line 14: | Line 18: |
| * Visualization tools, such as MRI visualization in 3 orthogonal views (either seperately or in 3D view), and overlayed surface visualization of cortex, skull, and scalp * Cortical surface extraction, using a multi-stage user friendly approach. Tools include brain surface extraction, bias field correction, voxel classification, cerebellum removal, and surface generation * Topological correction of cortical surfaces, which uses a graph-based approach to remove topological defects (handles and holes) and ensure a tessellation with spherical topology * Interactive labeling of sulcal landmarks in surface and orthogonal views * Skull and scalp surface extraction |
* [[http://brainsuite.org/|Brainsuite Documentation]] * [[http://brainsuite.org/download/|BrainSuite Downloads]] |
| Line 20: | Line 21: |
| A new version of BrainSuite will be released soon that will include coregistration of cortical surfaces to an atlas and surface-constrained volume registration to an atlas that produces a one-to-one mapping between cortical surfaces. | <<BR>><<BR>> |
BrainSuite
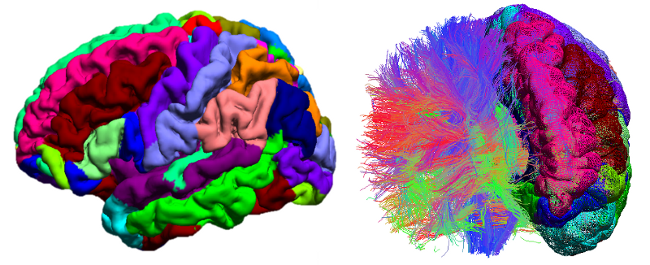
BrainSuite is a collection of image analysis tools designed to process magnetic resonance images (MRI) of the human head. BrainSuite13 provides an automatic sequence to extract cortical surface mesh models from the MRI, tools to register these to a labeled atlas to define anatomical regions of interest, and tools for processing diffusion imaging data. BrainSuite13 also contains visualization tools for exploring these data, and can produce interactive maps of regional connectivity.
BrainSuite is collaboratively developed between David Shattuck at the Ahmanson-Lovelace Brain Mapping Center at UCLA and Richard Leahy in the Biomedical Imaging Group, in collaboration with the Laboratory of Neuro Imaging, at USC. Other contributors to the development of the methods embodied in this software include: Anand A. Joshi, Chitresh Bhushan, Justin P. Haldar, Belma Dogdas, Stephanie Sandor-Leahy, Bijan Timsari, and Paul M. Thompson.
BrainSuite features include:
- Automated cortical surface extraction from T1-weighted MRI. Tools include brain surface extraction, bias field correction, voxel classification, cerebrum labeling, and surface generation
- Automated surface/volume registration software
- Tools for correction of B0 effects in diffusion data and coregistration to structural MRI
- Tools for processing of diffusion data including ODF and tensor fitting and tractography
- Sophisticated tools for visualizing and exploring MRI data, diffusion data, tractography, and connectivity
The currently released version of the software (BrainSuite) includes graphical user interface versions for Mac OSX and Windows and command line versions for Mac OSX, Windows and Linux. More details can be found on following links:
