|
Size: 26816
Comment:
|
Size: 17982
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 6: | Line 6: |
| == Current topics == ==== Source modeling ==== * Implementation of a new unified minimum norm/beamformer framework (almost finished) ==== Documentation ==== * Cross-validation with MNE and FieldTrip (work in progress): <<BR>>http://martinos.org/mne/dev/auto_tutorials/plot_brainstorm_auditory.html ==== Functional connectivity ==== * Significance thresholding of the connectivity matrices (not started) ==== Computation ==== * Removing the dependence to the Java interface to run in headless mode (not started) * Call MNE-Python and FieldTrip functions from Brainstorm GUI (work in progress) <<BR>><<BR>> |
|
| Line 24: | Line 7: |
| * RAW file viewer:<<BR>> | * Default montages for EEG (sensor selection) * Sleep scoring wish list (Emily C): * Configurable horizontal lines (for helping detecting visually some thresholds) * Mouse ruler: Measure duration and amplitude by dragging the mouse. * Automatic spindle detector * https://neuroimage.usc.edu/forums/t/page-overlap-while-reviewing-raw-file-a-way-to-set-to-0/11229/13 * RAW file viewer speed: |
| Line 29: | Line 18: |
| * Add field "comment" to markers: For clinicians to add notes (Marcel) * Events: Change the category of a selected event easily, instead of deleting/marking new * Events: Advanced process for recombining.<<BR>>Example: http://www.erpinfo.org/erplab/erplab-documentation/manual/Binlister.html * Bad trials: When changing the status of bad to good: remove the bad segments as well, otherwise it is not processed by processes like the PSD. * Review clinical recordings: Reduce the dimensionality of the data with a simple inverse problem, similar to what we do for the magnetic extrapolation ("Regional sources" in BESA, cf S Rampp) |
|
| Line 35: | Line 19: |
| * 2DLayout: * Overlay multiple conditions * Does not work when DC offset is not removed * Add a proper amplitude scale that gets updated when shift+scroll, to compare figures * Update the vertical lines when changing the gain (shift+scroll) |
* Create heat maps: Maybe with matlab function heatmap? * BioSemi: Add menu "Convert naming system" to rename channels into 10-10 (A1=>FPz) |
| Line 43: | Line 24: |
| * Start Brainstorm without Java (-nodesktop) * Option to increase the size of all the elements of the interface (for high res laptop screens) |
|
| Line 49: | Line 29: |
| * Create a colormap similar to MNE, where extrema are bright | |
| Line 51: | Line 30: |
| * Open new figures as tab (docked in the Figures window) | |
| Line 53: | Line 31: |
| * Removing all the CTRL and SHIFT in the keyboard shortcuts * Display warning before opening files that are too big * Smooth display from figure_image (ERPimage, raster plot...) |
|
| Line 59: | Line 34: |
| * Use boundary() instead of conhull() in all the display functions (ie. 2DDisc) | * Progress bar: Add a "Cancel" button * Error message: Add a link to report directly the bug on the forum * Reorganize menus (Dannie's suggestion): {{attachment:dannie_menus.png||width="382",height="237"}} |
| Line 62: | Line 40: |
| * Thresholding and stat tests the connectivity matrices | * Thresholding and stat tests for connectivity matrices |
| Line 64: | Line 42: |
| * Display of connectivity graphs: * Display as straight lines * Recode 2D graphs * 3D display with anatomical constrains * Use new band-pass filters in bst_connectivity ('bst-hfilter' instead of 'bst-fft-fir') * Review by Jan-Mathijs: http://journal.frontiersin.org/article/10.3389/fnsys.2015.00175/full * Connectivity based on band limited power (Sylvain): * Compute Hilbert/Bandpass + correlation of the envelopes * Bandpass envelopes before computing correlations? * Compute Hilbert(sensors) and then project to source space? * Graph view: * Does not display negative values correctly (correlation or difference of coherence) * Re-write using pure Matlab code and smoothed graphics * Fixed scales for intensity sliders * Text bigger * Too much data in appdata * Fixed scales for intensity sliders * Add "=" shortcut for having graphs with similar configurations * Disable zoom in one region (serious bugs) * NxN on sensors: does not place the sensors correctly in space * Coherence: * Average cross-spectra instead of concatenating epochs (to avoid discontinuities)<<BR>>Explore inter-trial approaches (Esther refers to chronux toolbox) * Granger: * Crashes sometimes: improve stability * Check for minimum time window (Esther: min around 500-1000 data points) * Re-write and optimize code * Add progress bar |
* Connect NxN display: * Graph on sensors: does not place the sensors correctly in space * Display as image: Add legend of the elements along X and Y axis * Display as time series: Display warning before trying to open too many signals * Time-resolved correlation/coherence: Display as time bands * Weighted Phase Lag Index (WPLI) * Coherence: Average cross-spectra instead of concatenating epochs (to avoid discontinuities)<<BR>>Explore inter-trial approaches (Esther refers to chronux toolbox) * Granger: Check for minimum time window (Esther: min around 500-1000 data points) |
| Line 92: | Line 51: |
| * Add p-values | |
| Line 94: | Line 52: |
| * Optimize code | |
| Line 98: | Line 55: |
| * PAC: * Add input TF , to disconnect TF decomposition and PAC computation (Peter) * Refine frequency vector of low frequencies * How many central frequencies to use in bst_pac? * Change filters: no chirplet functions * bst_freqfilter: Use nfcomponents like in bst_pac * Esther recommended a larger frequency binning of the PAC estimation * PAC maps: Display all sensors at once (like TF and DynamicPAC) * Hui-Ling's PAC: * https://bsp.hackpad.com/Cross-Frequency-Coupling-cChe95lhDHz * https://github.com/NCTU-BSP/MEEG * Time-resolved correlation/coherence: Display as time bands * Other metrics: * Coherence by bands: bst_coherence_band_welch.m * Granger by bands: bst_granger_band.m * Inter-trial coherence * Tutorial coherence [1xN] : Reproduce FieldTrip results? * Connect NxN: Display as time series > Display warning before trying to open too many signals |
|
| Line 118: | Line 57: |
| * Use CUDA for speeding up some operations (filtering, wavelets, etc) * Allow processes in Python and Java |
* Plugin manager: * Export all the software environment to a .zip file (brainstorm + all plugins) * Generate fully reproducible scripts, including all the interactive/graphical parts: * Saving all the interactive operations as process calls * Improving the pipeline editor to handle loops over data files or subjects * Keeping a better track of the provenance of all the data (History, uniform file names) |
| Line 121: | Line 64: |
| * scikit-learn classifiers: S Marti / G Dehaene * Implement data exchange with MNE-Python: write FIF files from Brainstorm and/or pass python objects in memory instead of FIF files * SSS/tSSS cleaning |
* scikit-learn classifiers * https://neuroimage.usc.edu/forums/t/ica-on-very-long-eeg/23556/4 * https://neuroimage.usc.edu/forums/t/best-way-to-export-to-mne-python/12704/3 |
| Line 125: | Line 68: |
| * Point-spread functions (PSFs) and cross-talk functions: https://mne.tools/stable/auto_examples/inverse/plot_psf_ctf_vertices.html#sphx-glr-auto-examples-inverse-plot-psf-ctf-vertices-py * Spatial resolution metrics in source space:<<BR>>https://mne.tools/stable/auto_examples/inverse/plot_resolution_metrics.html#sphx-glr-auto-examples-inverse-plot-resolution-metrics-py |
|
| Line 136: | Line 81: |
| * ft_prepare_sourcemodel: Compute MNI transformation (linear and non-linear) => Peter | * ft_prepare_heamodel: Add support from BEM surfaces from the Brainstorm database |
| Line 138: | Line 83: |
| * ft_read_atlas('TTatlas+tlrc.BRICK'); | |
| Line 142: | Line 86: |
| * Allow FieldTrip functions in compiled version * Decoding/Classifiers: Faster algorithms (MNE-Python?) |
* Optimization: * Use CUDA for speeding up some operations (filtering, wavelets, etc) * Use Matlab Coder to optimize: Wavelets, bandpass filter, sinusoid removal |
| Line 147: | Line 92: |
| * Events: Allow selection from a drop-down list (similar to option "channelname" in panel_process_selection) | |
| Line 150: | Line 96: |
| * Add Alex's suggestions: https://neuroimage.usc.edu/forums/t/ica-on-very-long-eeg/23556/4 * Add methods: SOBI, Fastica, AMICA/CUDICA/CUDAAMICA (recommended by S Makeig) |
|
| Line 152: | Line 100: |
| * Add a stand-alone tutorial | |
| Line 156: | Line 103: |
| * Use faster methods (MNE-Python?) * Add methods: SOBI, Fastica, AMICA/CUDICA (recommended by S Makeig) |
|
| Line 159: | Line 104: |
| * S Makeig: Use ICA to select the IC of interest instead of only removing artifacts * Display of spectrum for components (PSD/FFT) * Use FastICA (algo crashing) * Add components preselection: Correlation with EOG/ECG |
|
| Line 167: | Line 108: |
| * Save IC time series in database | * Use EYE-EEG: EEGLAB toolbox for eye-tracker guided ICA (Olaf Dimigen): http://www2.hu-berlin.de/eyetracking-eeg/ |
| Line 170: | Line 111: |
| * Show where the attenuation is projected:<<BR>>(sum(IK,2)-sum(SSP(k,:)*IK,2)./sum(IK,2) * Pipeline editor: * When computing sources from the pipeline editor: doesn't reselect the options if you click twice on "edit" (works for minnorm, but not for lcmv) * When computing time-frequency/hilbert/psd: Find a way not to force the user to click on Edit * Bandpass: * Use new filters in all the functions using a bandpass ('bst-hfilter' instead of 'bst-fft-fir'): process_evt_detect_threshold * Weird bug: Filter(import) != Import(Filter) in the HCP tutorial... to investigate * Bandpass * Spectral flattening (John): * ARIMA(5,0,1): Apply on the signal before any frequency/connectivity/PAC analysis * PSD: * Rewrite to have the same input as coherence (frequency resolution instead of window length) * Use the progress bar * Allow display of Avg+StdErr |
|
| Line 188: | Line 112: |
| * Use Matlab Coder to optimize some processes: Wavelets, bandpass filter, sinusoid removal * Reports: Click on link reopens exactly the figure |
|
| Line 193: | Line 115: |
| * Matching pursuit: http://m.jneurosci.org/content/36/12/3399.abstract?etoc * Bug: Display logs as negative * Bug: 3D figures: Colormaps with "log" option doesn't work * Bug: Difference of power displayed in log: problems (Soheila) * 2D Layout in spectrum * Make much faster and more memory efficient (C functions coded by Matti ?) * TF scouts: should display average of TF maps * Impossible to keep complex values for unconstrained sources * Pad short epochs with zero values for getting lower frequencies |
|
| Line 203: | Line 116: |
| * Extend clusters tab to display of TF to overlay TF signals (Svet) | |
| Line 208: | Line 120: |
| * Artifact detection: * Artifact rejection like SPM: if bad in 20%, bad everywhere * Test difference between adjacent samples * Events detection: Add option "std" vs "amplitude" * Co-registration of MEG runs: * SSP: Group projectors coming from different files * Finish validation of the method * Apply to continuous recordings for correcting head movements * Other processes: * Moving average * Max * Median * Simulation: * Fix units in simulation processes => no *1e-9 in "simulate recordings" * Use "add noise" process from Hui-Ling (in Work/Dev/Divers) * Use field process field "Group" to separate Input/Processing/Output options * Use new Matlab functions: movmean, movsum, movmedian, movmax, movmin, movvar, movstd * Process ft_prepare_heamodel: Add support from BEM surfaces from the Brainstorm database == Database == * Add buttons to sort files: by name, by comment, by date * MEG protocols: More flexible organization of the database; sub-conditions to allow different runs X different conditions. * Matrix files: Allow to be dependent from other files * Add notes in the folders (text files, visible as nodes in the tree) * Screen captures: save straight to the database * Rename multiple files * Default headmodel lost when reloaded: Keep selection on the hard drive (in brainstormstudy.mat) * protocol.mat can be too big: do not store the results links in it (and recreate when loading)- http://neuroimage.usc.edu/forums/t/abnormally-slow-behavior/2065/10 == Distributed computing == * Options from FieldTrip: * Loose collection of computers: https://github.com/fieldtrip/fieldtrip/tree/master/peer * Alternative, with less limitations: http://research.cs.wisc.edu/htcondor/ * Single multicore machine: https://github.com/fieldtrip/fieldtrip/tree/master/engine * Batch system: https://github.com/fieldtrip/fieldtrip/tree/master/qsub * Documentation: http://fieldtrip.fcdonders.nl/faq#distributed_computing_with_fieldtrip_and_matlab * PSOM: http://psom.simexp-lab.org/ * Various initiatives: http://samirdas.github.io/Data_sharing.html#/ |
|
| Line 253: | Line 122: |
| * Menu head model > Copy to other conditions/subjects (check if applicable first) * Menu Sources > Maximum value: Doesn't work with volume or mixed head models * Mixed head models: <<BR>> * Set loose parameter from the interface |
* Unconstrained sources: * Unconstrained to flat: Default PCA for stat and connectivity? * Process "Scouts time series": Add PCA option (replace isnorm with radio PCA/Norm) * Reproduce results in "Simultaneous human intracerebral stimulation and HD-EEG, ground-truth for source localization methods": https://www.nature.com/articles/s41597-020-0467-x * eLORETA instead of sLORETA? * https://neuroimage.usc.edu/forums/t/compute-eeg-sources-with-sloreta/13425/6 * "eLORETA algorithm is available in the MEG/EEG Toolbox of Hamburg (METH)": https://www.biorxiv.org/content/biorxiv/early/2019/10/17/809285.full.pdf * https://github.com/brainstorm-tools/brainstorm3/issues/114 * Sensitivity maps: https://mne.tools/stable/auto_examples/forward/plot_forward_sensitivity_maps.html * Point-spread and cross-talk functions (code in MNE-Python): * https://www.biorxiv.org/content/biorxiv/early/2019/06/18/672956.full.pdf * https://github.com/olafhauk/EEGMEGResolutionAtlas * Dipoles: * Project individual dipoles files on a template * panel_dipoles: Doesn't work with multiple figures * Project sources: Very poor algorithm to project sub-cortical regions and cerebellum * Mixed head models: Bug when displaying interpolated in MRI viewer * Maximum: * Menu Sources > Maximum value: Doesn't work with volume or mixed head models * Panel Get coordinates: Add button "find maximum" * Sources on surface: Display peak regions over time (time = color) => A.Gramfort * BEM single sphere: Get implementation from MNE |
| Line 260: | Line 148: |
| * Menu Sources > Simulate recordings: * Do not close the 3D figures after generating a new file * Add a process equivalent to this menu * Panel Get coordinates: Add button "find maximum" * Dipoles: * Project individual dipoles files on a template * panel_dipoles: Doesn't work with multiple figures * BEM single sphere: Get implementation from MNE * Visualize Beamformer results: * Read CTF SAM .svl * Display as layers in the MRI viewer * Unconstrained sources: * Stat and connectivity: what to do? (re-send email John+Sylvain) * Overlapping spheres: improve the estimation of the spheres for the frontal lobes * Magnetic extrapolation: Do the same thing with EEG * Noise covariance matrix: * Display with figure_image() * Storage of multiple noise covariance matrices (just like the head models) * Always save as full, then at inversion time, we can decide between full, heteroskedastic (diagonal) or homoskedastic (i.i.d, scalar) * Problem of having inividual trials + averages in the condition => Display warning or not? * Save nAvg in noisecov file, to make it easier to scale to other recordings * Sources on surface: Display peak regions over time (time = color) => A.Gramfort * Calculate ImagingKernel * Gain for a scout * Time-frequency beamformers: * Band-pass everything in different frequency bands + Source estimation + TF * Ask data to Sarang where he sees effects that cannot be extracted with MN followed by TF * Process "Extract scouts time series": Add PCA option (replace isnorm with choice PCA/Norm) * BEM: Fix unstable results when one vertex is too close from the layers (5mm ?) * Hui-Ling beamformers: * More explanations about what is in NAI and Spatial filters * Explain that is this is better to study effects extended in time (Ntime > Nsensors) * Group LCMV+MCB * Condition LEFT median nerve: very bad results * Keep options for inverse computation * OpenMEEG: Post example datasets for the remaining issues: * https://github.com/openmeeg/openmeeg/issues/64 * https://github.com/openmeeg/openmeeg/issues/68 * Example protocol ECOG: doesn't work |
|
| Line 299: | Line 149: |
| * Display spectrum scouts (PSD plots when clicking on "Display scouts" on PSD/full cortex) | |
| Line 301: | Line 152: |
| * Registration: * Use the same registration for multiple recording sessions that have already re-registered previously (eg. with MaxFilter) * Check templates: MNI transformation and volume atlases for ICBM152 vs Colin27 (loading the AAL atlas as surface or voume scouts do not align well on the ICBM152) * Compute non-linear MNI registration instead of linear * Select and remove bad digitized head points before automatic coregistration * Load the MNE -transf.fif: http://neuroimage.usc.edu/forums/showthread.php?2830 |
* FastSurfer: https://deep-mi.org/research/fastsurfer/ * '''SimNIBS''': Replace HEADRECO with CHARM (headreco will be removed in SimNIBS 4) |
| Line 311: | Line 159: |
| * Add keyboard shortcuts to scroll in the three orientations (same in MRI 3D) | |
| Line 313: | Line 160: |
| * Render surface envelope in the MRI as a thin line instead of the full interpolation matrix * Edit fiducials: Replace 6 text boxes with 1 for easy copy-paste (see fiducials.m) * Optimize computation interpolation MRI-surface (tess_tri_interp) => spm_mesh_to_grid * BrainVISA: Add support for MarsAtlas (Guillaume A) |
* Render surface envelope in the MRI as a thin line instead of the full interpolation matrix<<BR>>Or use inpolyhedron to get a surface mask and then erode it to get the volume envelope * Surface>Volume interpolation: Use '''spm_mesh_to_grid''' instead of tess_tri_interp |
| Line 318: | Line 163: |
| * Use BrainSuite inner skull for surface generation | |
| Line 320: | Line 164: |
| * Use for volume coregistration (rigid / non-rigid) * USCBrain: Add default electrodes positions * Templates for different ages: * Pediatric head atlases: https://www.pedeheadmod.net/pediatric-head-atlases-v1-2/ * https://iopscience.iop.org/article/10.1088/2057-1976/ab4c76 * https://www.biorxiv.org/content/biorxiv/early/2020/02/09/2020.02.07.939447.full.pdf * John Richards: https://www.nitrc.org/frs/?group_id=1361 * Neurodev database: https://jerlab.sc.edu/projects/neurodevelopmental-mri-database/ |
|
| Line 322: | Line 176: |
| * Display scouts in a tree: hemisphere, region, subregion * Sort scouts by region in process options * Downsample to atlas: allow on timefreq/connect files |
* Project from one hemisphere to the other using registered spheres/squares (http://neuroimage.usc.edu/forums/t/how-to-create-mirror-roi-in-the-other-hemisphere/5910/8) |
| Line 326: | Line 178: |
| * Major bug when importing surfaces for an MRI that was re-oriented manually * Smooth surface: Fix little spikes due to irregularities in the mesh * Bug: Hide scouts in the preview of the grid for volume head models |
* Geodesic distance calculations:<<BR>>https://www.mathworks.com/matlabcentral/fileexchange/6110-toolbox-fast-marching |
| Line 331: | Line 181: |
| * Contact positions: Import / set / detect<<BR>> * Automatic segmentation of CT scans: See project of Rodrigo Paz (Marseille/Timone), or check with V Litvak if there is anything usable in Lead-DBS. * Import from FreeSurfer/FreeView: Problem of difference between RAS and TkRegRAS: http://neuroimage.usc.edu/forums/showthread.php?1958-SEEG-electrodes-and-subject-s-anatomy-are-not-alligned * Add history: Save modifications and transformations applied to the channel files (Marcel) * ECOG: How to handle cases where not all the grid contacts are in the channel file? (Marcel) |
* Contact positions: Import / set / detect * New option: Align on none|inner|cortex to replace ECOG-mid |
| Line 337: | Line 184: |
| * Create clusters from anatomical labels: * Identify contacts in a given anatomical region (volume scout, surface mesh, or label in a volume atlas) / allow extracting the signals from all the contacts in an ROI * Automatic segmentation of CT: * GARDEL: http://meg.univ-amu.fr/wiki/GARDEL:presentation * SEEG DEETO Arnulfo 2015: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-015-0511-6 * Used routinely at Niguarda Hospital + other hospitals worldwide, reliable tool. * To be used with SEEG-assistant/3DSlicer: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-017-1545-8 * ECOG Centracchio 2021: https://link.springer.com/content/pdf/10.1007/s11548-021-02325-0.pdf * Classifier on thresholded CT: https://github.com/Jcentracchio/Automated-localization-of-ECoG-electrodes-in-CT-volumes * SEEG Granados 2018 (no code shared): https://link.springer.com/content/pdf/10.1007/s11548-018-1740-8.pdf * ECOG: * Project and display contacts on cortex surface should consider the rigidity of the grids: Contacts cannot rotate, and distance between contacts should remain constant across runs * Method for contacts projection: https://pdfs.semanticscholar.org/f10d/6b899d851f3c4b115404298d7b997cf1d5ab.pdf * ECOG: Brain shift: When creating contact positions on a post-implantation image, the brain shift should be taken into account for creating images of the ECOG contacts on the pre-op brain => iELVis (http://ielvis.pbworks.com/w/page/116347253/FrontPage) |
|
| Line 338: | Line 199: |
| * Display SEEG+ECOG contacts at the same time * Better way to represent ECOG strips that are not in contact with the skull (new type?) |
|
| Line 341: | Line 200: |
| * ECOG: Topography display without the 3D coordinates (Marcel) * 2DLayout: Should not try to use the actual position of the sensors, just the grids/strips. |
|
| Line 344: | Line 201: |
| * Alternatives to OpenMEEG: SimBio/FieldTrip? Matti Stenroos? NFT/NIST? * Prepare tutorial with public dataset (Marcel) |
* Display curved SEEG electrodes * Detection CEEP stim artifacts: Use ImaGIN code ImaGIN_StimDetect |
| Line 348: | Line 205: |
| * Stat on connectivity? * Stat on unconstrained sources? |
|
| Line 349: | Line 208: |
| * Use LENA functions(?) | * Write panel similar to Process1 and Process2 |
| Line 353: | Line 212: |
| * New process to test for Gaussianity using swtest * PLS: Partial Least Squares * Simulate recordings with specific properties, for stat validation * Quality control before statistics, on condition averages across subjects:<<BR>>mean(baseline)/std(baseline): shows bad subject quickly. * Use SurfStat: Impements interesting things, like an analytical cluster-based p-value correction (Random-field theory which is used in SPM) - Peter |
|
| Line 360: | Line 213: |
| * '''XDF import''': Use the EEGLAB plugin, contact Martin Bleichner (Oldenburg) * BIDS import: For CTF files, the head shape might be in the session folder instead of the .ds |
* BIDS import: * Read real fiducials (OMEGA) / transformation matrices: * https://groups.google.com/g/bids-discussion/c/BeyUeuNGl7I * https://github.com/bids-standard/bids-specification/issues/752#issuecomment-795880992 * Read associated empty room * BIDS export: * Add events tsv, channel tsv, EEG, iEEG * BIDS-Matlab? * Support for OpenJData / JNIfTI: https://github.com/brainstorm-tools/brainstorm3/issues/284 |
| Line 365: | Line 225: |
| * FieldTrip: Import/Export time-frequency: * Export: http://neuroimage.usc.edu/forums/t/export-time-frequency-to-fieldtrip/1968 * Import: http://neuroimage.usc.edu/forums/t/import-time-frequency-data-from-fieldtrip/2644 |
* SPM .mat/.dat: Fix the import of the EEG/SEEG coordinates * EEG File formats: * XDF: https://github.com/sccn/xdf * XLTEK: https://github.com/danielmhanover/OpenXLT * Persyst .lay: https://github.com/ieeg-portal/Persyst-Reader * Nervus .eeg: https://github.com/ieeg-portal/Nervus-Reader * Biopac .acq: https://github.com/ieeg-portal/Biopac-Reader * BCI2000 Input (via EEGLAB plugin) |
| Line 369: | Line 235: |
| * Use reader from MNE-Python: mne.io.read_raw_kit (doesn't require Yokogawa slow library) | * Use reader from MNE-Python: mne.io.read_raw_kit (skip Yokogawa slow library) |
| Line 373: | Line 239: |
| * References at too far from the head sensors in Marseille 4D system | |
| Line 376: | Line 241: |
| * EEG File formats:<<BR>> * Nicolet EEG: Get from from MNE * EEG CeeGraph * EGI: Finish support for epoched files (formats 3,5,7) * BCI2000 Input (via EEGLAB plugin) * EEGLAB import: * Support for binary AND epoched files (now it's one or the other) * Allow epoched files with recordings saved in external files * BST-BIN: Add compressionto .bst * XLTEK: https://github.com/danielmhanover/OpenXLT * Persyst .lay: https://github.com/ieeg-portal/Persyst-Reader * Nervus .eeg: https://github.com/ieeg-portal/Nervus-Reader * Biopac .acq: https://github.com/ieeg-portal/Biopac-Reader * Review raw on all the file formats (ASCII EEG and Cartool missing) * gTec EEG recordings: Read directly from the HDF5 files instead of the Matlab exports. * Use new Matlab functions readtable/writetable (2014a): for Excel and text files * SPM .mat/.dat: Fix the import of the EEG/SEEG coordinates |
* BST-BIN: Add compression to .bst |
| Line 395: | Line 244: |
| * Add tags to the forum posts for easier listing by topic | * Count GitHub clones in the the download stats * Deface the MRIs of all the tutorials |
| Line 397: | Line 247: |
| * Update the organization of derivatives folder (same for ECOG tutorial) | |
| Line 399: | Line 250: |
| * New tutorials: * Visual group Henson/Wakeman tutorial: Use the BIDS version<<BR>>https://github.com/INCF/BIDS-examples/tree/bep008_meg/ds000117 |
* New tutorials: <<BR>> |
| Line 402: | Line 252: |
| * Rat PAC + high gamma (Soheila) | |
| Line 405: | Line 254: |
| * FieldTrip cortico-muscular coherence tutorial: http://www.fieldtriptoolbox.org/tutorial/coherence | |
| Line 408: | Line 256: |
* MEG steady-state / high-gamma visual / frequency tagging * BIDS-EEG example datasets * Intra-cranial recordings (Marcel, Jeremy?) * Stand-alone ICA tutorial * Co-register MEG runs (Beth) * Move all the files to download on the cloud for faster download everywhere in the world * Provide secure way of sending password over HTTPS for: * Account creation * Forum exchanges * org.brainstorm.dialog.CloneControl * Workflows FieldTrip: http://www.fieldtriptoolbox.org/faq/what_types_of_datasets_and_their_respective_analyses_are_used_on_fieldtrip * Reference tutorials on Google scholar + ResearchGate * Count GitHub clones in the the download stats * Google Analytics: Create template and update the section of the Community page * Missing in page "Cite Brainstorm": Add all the methods used in the software * Deface the MRIs of all the tutorials * Compiled R2016b: Color picker doesn't work (for changing surface color for instance) |
* MEG steady-state / high-gamma visual / frequency tagging * Reproduce results from "Simultaneous human intracerebral stimulation and HD-EEG, ground-truth for source localization methods": https://www.nature.com/articles/s41597-020-0467-x * Stand-alone ICA tutorial |
| Line 430: | Line 261: |
| * Screen capture: * Bug on Win8/Win10: doesn't capture the correct part of the screen * Window managers with fading effect: captures the top window |
|
| Line 442: | Line 270: |
| * MacOS 10.14.5 (Mojave): * Toggle buttons do not show their status * Panel Record: Text is too large for text boxes |
|
| Line 446: | Line 277: |
| * Matlab bugs: * Interface looks small on screens with very high resolutions: Reduce the resolution * Event markers are not visible anymore with the sequence: Open MEG, open EOG, close MEG * in_bst_data_multi: If trials have different sizes, output is random (the one of the first file) * Edit scout in MRI: small modifications cause huge increase of the scout size * Canolty maps computation: Fix progress bar |
== Distributed computing == * Options from FieldTrip: * Loose collection of computers: https://github.com/fieldtrip/fieldtrip/tree/master/peer * Single multicore machine: https://github.com/fieldtrip/fieldtrip/tree/master/engine * Batch system: https://github.com/fieldtrip/fieldtrip/tree/master/qsub * Documentation: https://www.fieldtriptoolbox.org/faq/what_are_the_different_approaches_i_can_take_for_distributed_computing/ * PSOM: http://psom.simexp-lab.org/ |
| Line 454: | Line 287: |
| * bst_bsxfun: After 2016b, we can use directly the scalar operators (./ .* ...) instead of bsxfun. Update bst_bsxfun to skip the use of bsxfun when possible. | * Replace all calls to inpolyhd.m with inpolyhedron.m (10x faster) |
| Line 456: | Line 289: |
| * Hide Java panels instead of deleting them | |
| Line 459: | Line 291: |
| * bst_warp and channel_project: Use tess_parametrize_new instead of tess_parametrize * Shared kernels: "get bad channels" operation in a different way (reading all the files is too slow) * Optimize bst_get: * Now study and subject have necessarily the same folder name * Replace big switch with separate functions * Progress bar: * Add different levels (to handle sub-processes) * Make work correctly with RAW on resting tutorial * Uniformize calls in bst_process/Run * Add a "Cancel" button * Fix all the 'todo' blocks in the code * Error message: Add a link to report directly the bug on the forum |
What's next
A roadmap to the future developments of Brainstorm.
Contents
Recordings
- Default montages for EEG (sensor selection)
- Sleep scoring wish list (Emily C):
- Configurable horizontal lines (for helping detecting visually some thresholds)
- Mouse ruler: Measure duration and amplitude by dragging the mouse.
- Automatic spindle detector
https://neuroimage.usc.edu/forums/t/page-overlap-while-reviewing-raw-file-a-way-to-set-to-0/11229/13
- RAW file viewer speed:
- Downsample before filtering? (attention to the filter design)
- Add parameter to make the visual downsampling more or less aggressive
- Pre-load next page of recordings
- Keep the filter specifications in memory instead of recomputing for every page
- MEG/EEG registration: Apply the same transformation to multiple runs
- Create heat maps: Maybe with matlab function heatmap?
BioSemi: Add menu "Convert naming system" to rename channels into 10-10 (A1=>FPz)
Interface
Add a warning when computing a forward model with > 100000 sources (check selection)
- Snapshot: Save as image / all figures (similar to Movie/all figure)
Generalize the use of the units (field .DisplayUnits): Rewrite processes to save the units correctly
- Colormaps:
- Allow brightness/contrast manipulations on the custom colormaps
- Global colormap max: Should get the maximum across all the open files
- Copy figures to clipboard (with the screencapture function)
Contact sheets & movies: use average of time windows instead of single instants, for each picture.
- Contact sheets: Allow explicit list of times in input (+ display as in MNE-Python with TS)
- Display CTF coils: Show discs instead of squares
- Progress bar: Add a "Cancel" button
- Error message: Add a link to report directly the bug on the forum
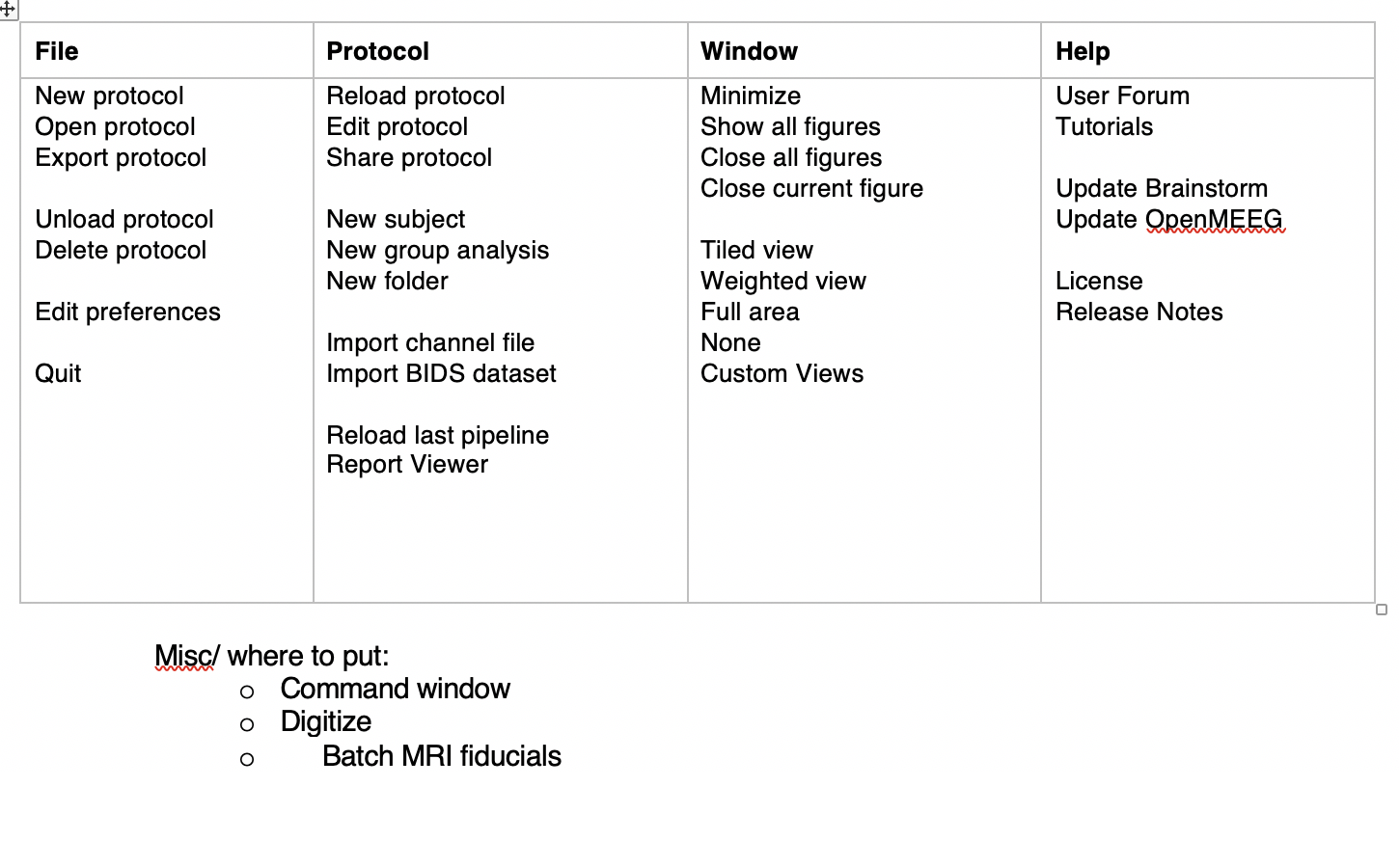
Reorganize menus (Dannie's suggestion):
Connectivity
- Thresholding and stat tests for connectivity matrices
- Connectivity on unconstrained sources: "Default signal extraction for volume grids should be the time series of the first principal component of the triplet signals after each has been zero-meaned" (SB)
- Connect NxN display:
- Graph on sensors: does not place the sensors correctly in space
- Display as image: Add legend of the elements along X and Y axis
- Display as time series: Display warning before trying to open too many signals
- Time-resolved correlation/coherence: Display as time bands
- Weighted Phase Lag Index (WPLI)
Coherence: Average cross-spectra instead of concatenating epochs (to avoid discontinuities)
Explore inter-trial approaches (Esther refers to chronux toolbox)- Granger: Check for minimum time window (Esther: min around 500-1000 data points)
- PLV:
- Remove evoked
- Add time integration
- Unconstrained sources
- Add warning when running of short windows (because of filters)
Processes
- Plugin manager:
- Export all the software environment to a .zip file (brainstorm + all plugins)
- Generate fully reproducible scripts, including all the interactive/graphical parts:
- Saving all the interactive operations as process calls
- Improving the pipeline editor to handle loops over data files or subjects
- Keeping a better track of the provenance of all the data (History, uniform file names)
- Add MNE-Python functions:
- scikit-learn classifiers
https://neuroimage.usc.edu/forums/t/ica-on-very-long-eeg/23556/4
https://neuroimage.usc.edu/forums/t/best-way-to-export-to-mne-python/12704/3
- Reproduce other tutorials / examples
Point-spread functions (PSFs) and cross-talk functions: https://mne.tools/stable/auto_examples/inverse/plot_psf_ctf_vertices.html#sphx-glr-auto-examples-inverse-plot-psf-ctf-vertices-py
Spatial resolution metrics in source space:
https://mne.tools/stable/auto_examples/inverse/plot_resolution_metrics.html#sphx-glr-auto-examples-inverse-plot-resolution-metrics-py- Change the graphic renderer from Matlab
Add FieldTrip functions:
- ft_sourceanalysis:
- Check noise covariance
- Check all the options of all the methods
- Single trial reconstructions + noise covariance?
Filters?? http://www.fieldtriptoolbox.org/example/common_filters_in_beamforming
Beamformers: Save ftSource.avg.mom
http://www.fieldtriptoolbox.org/workshop/meg-uk-2015/fieldtrip-beamformer-demohttp://www.fieldtriptoolbox.org/tutorial/beamformingextended
- Baseline? Two inputs?
- ft_prepare_heamodel: Add support from BEM surfaces from the Brainstorm database
- Freqanalysis: ITC
ft_volumereslice: http://www.fieldtriptoolbox.org/faq/how_change_mri_orientation_size_fov
- ft_freqanalysis
- ft_combineplanar
- ft_sourceanalysis:
- Optimization:
- Use CUDA for speeding up some operations (filtering, wavelets, etc)
- Use Matlab Coder to optimize: Wavelets, bandpass filter, sinusoid removal
- Pipeline editor:
- Bug: After "convert to continuous", the time of the following processes should change
- Add loops over subjects/conditions/trial groups
- Events: Allow selection from a drop-down list (similar to option "channelname" in panel_process_selection)
ITC: Inter-trial coherence (see MNE reports for group tutorial)
http://www.sciencedirect.com/science/article/pii/S1053811916304232- ICA:
Add Alex's suggestions: https://neuroimage.usc.edu/forums/t/ica-on-very-long-eeg/23556/4
- Add methods: SOBI, Fastica, AMICA/CUDICA/CUDAAMICA (recommended by S Makeig)
- Why doesn't the ICA process converge when using 25 components in the EEG tutorial?
- Add an option to resample the signals before computing the ICA decomposition
- Exploration: Add window with spectral decomposition (useful for muscle artifacts)
- Export IC time series (and then compute their spectrum): solves the problem above
Comparison JADE/Infomax:
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0030135Dimension reduction with PCA adds artifacts: Not done by default in EEGLAB
Contact: Stephen Shall Jones ( shall-jones@infoscience.otago.ac.nz )
Student Carl Leichter detailed this in his thesis- Import ICA matrices available in EEGLAB .set files
EEGLAB recommends ICA + trial rejection + ICA again: Impossible right now with Brainstorm
(http://sccn.ucsd.edu/wiki/Chapter_09:_Decomposing_Data_Using_ICA)ICA+machine learning: https://www.ncbi.nlm.nih.gov/pubmed/28497769
Automated artifact rejection: https://arxiv.org/abs/1612.08194
Use EYE-EEG: EEGLAB toolbox for eye-tracker guided ICA (Olaf Dimigen): http://www2.hu-berlin.de/eyetracking-eeg/
- SSP:
Display warning if changing the ChannelFlag while there is a Projector applied
Remove line noise: http://www.nitrc.org/projects/cleanline
- Time-frequency:
- Optimization: bst_timefreq (around l.136), remove evoked in source space: Average should be computed in sensor space instead of source space (requested by Dimitrios)
Short-time Fourier transform: http://www.mikexcohen.com/lectures.html
- Hilbert with time bands very slow on very long files (eg. 3600s at 1000Hz) because the time vector is still full (10^7 values): save compressed time vector instead.
- When normalizing with baseline: Propagate with the edge effects marked in TFmask
- Allow running TF on montages
- Review continuous files in time-frequency space (for epilepsy)
- Bug when computing TF on constrained and unconstrained scouts at the same time (in mixed head models for instance): uses only the constrained information and doesn't sum the 3 orientations for the unconstrained regions.
Source modeling
- Unconstrained sources:
- Unconstrained to flat: Default PCA for stat and connectivity?
- Process "Scouts time series": Add PCA option (replace isnorm with radio PCA/Norm)
Reproduce results in "Simultaneous human intracerebral stimulation and HD-EEG, ground-truth for source localization methods": https://www.nature.com/articles/s41597-020-0467-x
- eLORETA instead of sLORETA?
https://neuroimage.usc.edu/forums/t/compute-eeg-sources-with-sloreta/13425/6
"eLORETA algorithm is available in the MEG/EEG Toolbox of Hamburg (METH)": https://www.biorxiv.org/content/biorxiv/early/2019/10/17/809285.full.pdf
Sensitivity maps: https://mne.tools/stable/auto_examples/forward/plot_forward_sensitivity_maps.html
- Point-spread and cross-talk functions (code in MNE-Python):
- Dipoles:
- Project individual dipoles files on a template
- panel_dipoles: Doesn't work with multiple figures
- Project sources: Very poor algorithm to project sub-cortical regions and cerebellum
- Mixed head models: Bug when displaying interpolated in MRI viewer
- Maximum:
Menu Sources > Maximum value: Doesn't work with volume or mixed head models
- Panel Get coordinates: Add button "find maximum"
Sources on surface: Display peak regions over time (time = color) => A.Gramfort
- BEM single sphere: Get implementation from MNE
- Volume grid:
- Optimize: 3D display (better than 9x9 cubes)
- Optimize: vol_dilate (with 26 neighbors)
- Add eyes models to attract eye activity
- Display spectrum scouts (PSD plots when clicking on "Display scouts" on PSD/full cortex)
Anatomy
FastSurfer: https://deep-mi.org/research/fastsurfer/
SimNIBS: Replace HEADRECO with CHARM (headreco will be removed in SimNIBS 4)
- MRI Viewer:
- Pan in zoomed view (shift + click + move?)
- Zoom in/out with mouse (shift + scroll?)
- Ruler tool to measure distances
- Display scouts as additional volumes
Render surface envelope in the MRI as a thin line instead of the full interpolation matrix
Or use inpolyhedron to get a surface mask and then erode it to get the volume envelopeSurface>Volume interpolation: Use spm_mesh_to_grid instead of tess_tri_interp
BrainSuite:
- Use same colors for left and right for anatomical atlases
- Use for volume coregistration (rigid / non-rigid)
- USCBrain: Add default electrodes positions
- Templates for different ages:
Pediatric head atlases: https://www.pedeheadmod.net/pediatric-head-atlases-v1-2/
https://www.biorxiv.org/content/biorxiv/early/2020/02/09/2020.02.07.939447.full.pdf
John Richards: https://www.nitrc.org/frs/?group_id=1361
Neurodev database: https://jerlab.sc.edu/projects/neurodevelopmental-mri-database/
- Scouts:
- Display edges in the middle of the faces instead of the vertices
Project from one hemisphere to the other using registered spheres/squares (http://neuroimage.usc.edu/forums/t/how-to-create-mirror-roi-in-the-other-hemisphere/5910/8)
- Parcellating volume grids: scikit-learn.cluster.Ward
Geodesic distance calculations:
https://www.mathworks.com/matlabcentral/fileexchange/6110-toolbox-fast-marching
ECOG/SEEG
- Contact positions: Import / set / detect
- New option: Align on none|inner|cortex to replace ECOG-mid
- Project contact positions across subjects or templates (Marcel)
- Create clusters from anatomical labels:
- Identify contacts in a given anatomical region (volume scout, surface mesh, or label in a volume atlas) / allow extracting the signals from all the contacts in an ROI
- Automatic segmentation of CT:
SEEG DEETO Arnulfo 2015: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-015-0511-6
- Used routinely at Niguarda Hospital + other hospitals worldwide, reliable tool.
To be used with SEEG-assistant/3DSlicer: https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-017-1545-8
ECOG Centracchio 2021: https://link.springer.com/content/pdf/10.1007/s11548-021-02325-0.pdf
Classifier on thresholded CT: https://github.com/Jcentracchio/Automated-localization-of-ECoG-electrodes-in-CT-volumes
SEEG Granados 2018 (no code shared): https://link.springer.com/content/pdf/10.1007/s11548-018-1740-8.pdf
- ECOG:
- Project and display contacts on cortex surface should consider the rigidity of the grids: Contacts cannot rotate, and distance between contacts should remain constant across runs
Method for contacts projection: https://pdfs.semanticscholar.org/f10d/6b899d851f3c4b115404298d7b997cf1d5ab.pdf
ECOG: Brain shift: When creating contact positions on a post-implantation image, the brain shift should be taken into account for creating images of the ECOG contacts on the pre-op brain => iELVis (http://ielvis.pbworks.com/w/page/116347253/FrontPage)
- Display:
- Bad channels: Contacts greyed out instead of ignored (Marcel)
- Display time in H:M:S
- Display curved SEEG electrodes
Detection CEEP stim artifacts: Use ImaGIN code ImaGIN_StimDetect
Statistics
- Stat on connectivity?
- Stat on unconstrained sources?
- ANOVA:
- Write panel similar to Process1 and Process2
- Output = 1 file per effect, all grouped in a node "ANOVA"
- Display several ANOVA maps (from several files) on one single figure, using a "graphic accumulator", towards which one can send any type of graphic object
Input / output
- BIDS import:
- Read real fiducials (OMEGA) / transformation matrices:
- Read associated empty room
- BIDS export:
- Add events tsv, channel tsv, EEG, iEEG
- BIDS-Matlab?
Support for OpenJData / JNIfTI: https://github.com/brainstorm-tools/brainstorm3/issues/284
- DICOM converter:
- Add dcm2nii (MRICron)
- Add MRIConvert
- SPM .mat/.dat: Fix the import of the EEG/SEEG coordinates
- EEG File formats:
Persyst .lay: https://github.com/ieeg-portal/Persyst-Reader
Nervus .eeg: https://github.com/ieeg-portal/Nervus-Reader
Biopac .acq: https://github.com/ieeg-portal/Biopac-Reader
- BCI2000 Input (via EEGLAB plugin)
- 4D file format:
- Use reader from MNE-Python: mne.io.read_raw_kit (skip Yokogawa slow library)
- Reference gradiometers: Keep the orientation of the first or second coil?
- Reference gradiometers: Add the sensor definition from coil_def.dat
- Validate with phantom recordings that noise compensation is properly taken into account
- The noise compensation is considered to be always applied on the recordings, not sure this assumption is always correct
- 4D phantom tutorial (JM Badier?)
- BST-BIN: Add compression to .bst
Distribution & documentation
Count GitHub clones in the the download stats
- Deface the MRIs of all the tutorials
- Tutorial OMEGA/BIDS:
- Update the organization of derivatives folder (same for ECOG tutorial)
- Add review of literature for the resting state MEG
- Download example datasets directly from the OMEGA repository
New tutorials:
Other public datasets: https://github.com/INCF/BIDS-examples/tree/bep008_meg/
- EEG/research
FieldTrip ECOG tutorial: http://www.fieldtriptoolbox.org/tutorial/human_ecog
Reproduce tutorials from MNE-Python: https://martinos.org/mne/stable/tutorials.html
Cam-CAN database: https://camcan-archive.mrc-cbu.cam.ac.uk/dataaccess/<<BR>>(download new datasets, including maxfiltered files and manual fiducial placements)
- MEG steady-state / high-gamma visual / frequency tagging
Reproduce results from "Simultaneous human intracerebral stimulation and HD-EEG, ground-truth for source localization methods": https://www.nature.com/articles/s41597-020-0467-x
- Stand-alone ICA tutorial
Current bugs
- Image viewer:
- Difficult to get to 100%
- Buggy on some systems
- 2DLayout:
- (TF) Units are weird with % values
- (TF) Difficult to navigate in frequencies: Scaling+changing frequency resets the scaling
- Progress bar:
- Doesn't close properly on some Linux systems
- Focus requests change workspace when processing constantly (Linux systems)
- MacOS 10.14.5 (Mojave):
- Toggle buttons do not show their status
- Panel Record: Text is too large for text boxes
- MacOS bugs:
- Buttons {Yes,No,Cancel} listed backwards
- Record tab: Text of epoch number is too big
- Colormap menus: Do not work well on compiled MacOSX 10.9.5 and 10.10
Distributed computing
Options from FieldTrip:
Loose collection of computers: https://github.com/fieldtrip/fieldtrip/tree/master/peer
Single multicore machine: https://github.com/fieldtrip/fieldtrip/tree/master/engine
Batch system: https://github.com/fieldtrip/fieldtrip/tree/master/qsub
Documentation: https://www.fieldtriptoolbox.org/faq/what_are_the_different_approaches_i_can_take_for_distributed_computing/
Geeky programming details
- Replace all calls to inpolyhd.m with inpolyhedron.m (10x faster)
Interface scaling: Rewrite class IconLoader to scale only once the icons at startup instead of at each request of an icon (might improve the speed of the rendering of the tree)
- Processes with "radio" and "radio_line" options: Replace with "radio_label" and "radio_linelabel"
- Interpolations: Use scatteredInterpolant, griddedInterpolant, triangulation.nearestNeighbor (2014b)
