Using the anatomy templates
Author: Francois Tadel, Raymundo Cassani
Brainstorm orients most of its database organization and processing stream for handling anatomical information together with the MEG/EEG recordings. The introduction tutorials start with the import of the T1 MRI of the subject, and this anatomy seems mandatory everywhere. These choices were made because the primary focus of Brainstorm was to estimate brain sources from MEG/EEG, which ideally requires an accurate spatial modelling of the head.
If you don't have access to anatomical images of your subjects or if you are not interested in source reconstruction, Brainstorm will still require that you explicitly define an anatomy; in those case you would use an anatomy template. In the case of group analysis at the source level, you would also use a template on which you would project of the individual results.
Several templates are available, with a large preference for using the MNI ICBM152 package distributed with Brainstorm because it provides the highest level of compatibility between different features within Brainstorm and with other software environments. You might be interested in using another one if you are working with different age ranges, or if you need to obtain results in a specific space. This tutorial provides references to the various templates available in Brainstorm.
Contents
Scenarios
An anatomy template is a set of anatomical files (MRI, surfaces, parcellations) representing a brain atlas. As the reference anatomy, we recommend using the ICBM125 2009c Nonlinear Asymmetric, distributed by default with Brainstorm, but other options are described below. An anatomy template can be used for various purposes:
Replacing the subject anatomy in the Brainstorm database when no individual scans are available for the subject, or when their quality is too low to process them with FreeSurfer or CAT. Setting the anatomy of the subject is necessary for 2D/3D topography display and source estimation.
Warping an anatomy template to match the digitized head shape of a subject. See tutorial: Warping the anatomy templates
For group analysis at the source level: an intermediate step of projection on an anatomy template is necessary. See tutorial: Group analysis: Subjects coregistration
When using an anatomy template as a substitution for the subject anatomy, you have to be careful with the spatial distortions it creates. As the reference anatomy has a different shape and cortical folding from your subject, with sometimes important differences in head size, it is preferable to follow these guidelines:
- If you have a head shape for the anatomy: Warp the anatomy.
- Chose a template close to the age range of the subject, especially for younger populations.
EEG: If you are not using default electrode positions distributed with the a Brainstorm template, align and project the electrodes on the template head (right-click on the channel file > MRI registration > Edit)
MEG: Using an anatomy template with MEG recordings is less indicated than with EEG, because it is not possible to move the MEG sensors to follow the head shape. Use this option with caution, and make sure the template you use has a similar head size as your subject. If you use the ICBM152 template to estimate brain sources from MEG recordings of a 1yr-old baby, most of your source space would be outside of the actual head of the baby.
Subject configuration
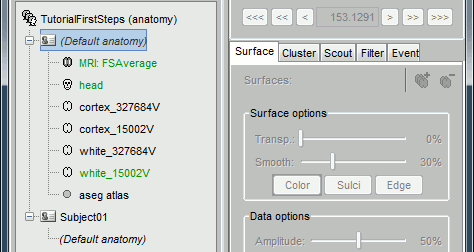
Using the default anatomy
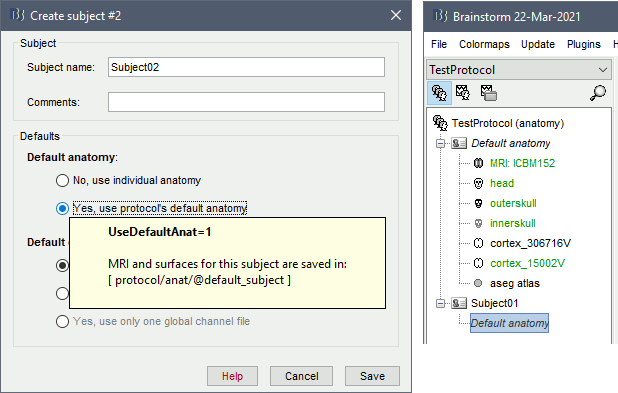
When creating a new subject, select the option "Yes, use protocol's default anatomy". This causes the contents of the subject anatomy folder to be replaced with the text Default anatomy. When a Brainstorm process or display function requests the default MRI or surfaces for this subject, the files from the Default anatomy are returned.
By default, all new protocols use as their default anatomy the simplified version of the ICBM152 MNI template distributed with Brainstorm (brainstorm3/defaults/anatomy/ICBM152). When creating the protocol, Brainstorm makes a copy of the ICBM152 anatomy, processed with FreeSurfer 6, and sets it as the default for the protocol. This template brain is used as a substitute for the subjects without an individual MRI, or as the common brain for group analysis.
Modify the default fiducials
The fiducial points (Nasion, LPA, RPA) used in your recordings might not be the same as the ones used in the anatomy templates in Brainstorm. By default, the LPA/RPA points are defined at the junction between the tragus and the helix, as represented with the red dot in the Coordinates systems page.
If you want to use an anatomy template but you are using a different convention when digitizing the position of these points, you have to modify the default positions of the template with the MRI Viewer.
- Go to the anatomy view
In (default anatomy), right-click on the MRI > Edit MRI
- Modify the position of the fiducial points to match your own convention
- Click on [Save], it will update the surfaces to match the new coordinate system
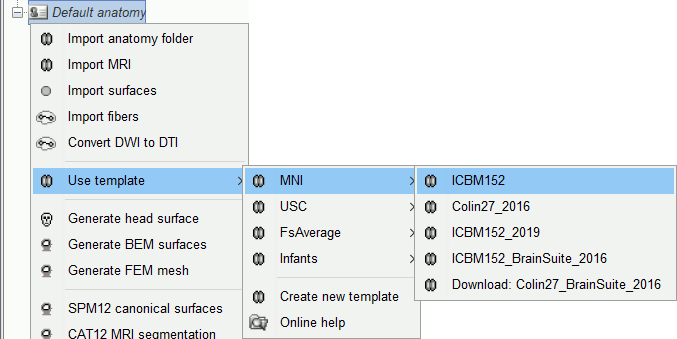
Changing the default anatomy
Other sets of MRI+surfaces are available to replace the ICBM152/FreeSurfer anatomy. Right-click on (Default anatomy) > Use template. If a package is not currently available on your system, it will be downloaded from the Brainstorm website and saved in $HOME/.brainstorm/defaults/anatomy.
If you click on any of the download options, it downloads it into your templates folder, then the list of files in the (default anatomy) folder is replaced with the new template.
If the automatic download doesn't work, you can download the templates manually from the Download page and copy the .zip files directly in the folder $HOME/.brainstorm/defaults/anatomy. More instructions in the Installation page.
Group analysis
When performing a group analysis with multiple subjects for which you have the individual MRI scans, you need to project the sources estimated on each subject on a common template, as explained in this tutorial: Group analysis.
For accurate registration between different brains (from a subject to a template or between subjects), you need to use a template that was generated using the same program as the one you used for running the segmentation of all the subjects of your studies.
You can use either BrainSuite or FreeSurfer/CAT12 for processing the MRIs or your subjects, but you need to use a template that matches this choice in order to use the accurate registration methods.
FreeSurfer templates
Available options:
ICBM152: Distributed directly with the Brainstorm package. Same as ICBM152_2023b, but without the white matter and mid surfaces, in order to minimize the size of the Brainstorm installation files.
ICBM152_2023b: Same as ICBM152_2023 but with updated BEM surfaces computed from FieldTrip, and cortex surfaces slightly modified not to cause any problem with OpenMEEG.
ICBM152_2023: ICBM 2009c Nonlinear Asymmetric, FreeSurfer 7.3.2, T1 only: more info
New volume atlases from FreeSurfer (7.3.2): ASEG, DKT, Deskian-Killiany, Destrieux.
New volume atlases from CAT12 (r2170): AAL3, Anatomy3, Cobra, Hammers, JulichBrain, LPBA40, Neuromorphometrics, Thalamus
- New tissue classification from SimNIBS4/CHARM: Tissues
- New surface atlases: Brainnetome, HCP-MMP1, PALS-B12, Schaefer, OASIS
- New features: Contralateral registration
ICBM152_2022: Same as ICBM152_2023, but without the volume parcellations from CAT12.
The template was generated with this package (scripts+data): ICBM152_2022_src.zip
ICBM152_2019: ICBM 2009c Nonlinear Asymmetric, FreeSurfer 6: more info
Colin27_2016: Average of 27 scans of the same head, FreeSurfer 5.3: more info
FsAverage_2020: Average of 40 subjects using a spherical averaging (Fischl et al. 1999).
It is the default FreeSurfer brain: please register here if you are using it.
More images: http://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer#FSAverage_templateOreilly infant templates: 13 anatomical models for subjects between zero and 24 months of age (O'Reilly et al. 2020)
They all include the following information:
- T1 MRI volume
- Cortex/white surface: high-resolution (~300.000 vertices) and low-resolution (15.000 vertices)
- Head layers: scalp, outer skull, inner skull
FreeSurfer spherical registration of each hemisphere, for subject co-registration.
FreeSurfer surface-based atlases: Desikan-Killiany, Destrieux, Brodmann, Mindboggle
(plus Yeo2011 and PALS for FSAverage only)- ASEG sub-cortical atlas.
For more information on the interactions between FreeSurfer and Brainstorm: read this tutorial.
BrainSuite templates
Available options:
Colin27_BrainSuite_2016: Average of 27 scans, processed with BrainSuite 15b: more information
ICBM152_BrainSuite_2016: Non-linear average of 152 subjects, BrainSuite 15b: more information
BCI-DNI_BrainSuite_2020: Single subject atlas from USC, BrainSuite 15c: more information
USCBrain_BrainSuite_2020: Anatomical and functional hybrid atlas from USC, BrainSuite 17a: more
Warning: If you are using BrainSuiteAtlas1 for BrainSuite processing, then you should use Colin27_BrainSuite_2016 or ICBM152_BrainSuite_2016 as the default anatomy. If you are using the BCI-DNI_brain_atlas, then you should use BCI-DNI_BrainSuite_2016 as the template in BrainStorm.
They all include the following information:
- T1 MRI volume
- Cortex/white surfaces: high-resolution (~300.000 vertices) and low-resolution (15.000 vertices)
- Head layers: scalp, outer skull, inner skull
BrainSuite square registration of each hemisphere, for subject co-registration.
BrainSuite surface-based atlas: SVReg
For more information on the interactions between BrainSuite and Brainstorm: read this tutorial.
BrainVISA templates
Note that the BrainVISA-based templates do not allow any accurate registration procedure.
Kabdebon_7w: 7-week infant brain with the anatomical atlas from (Kabdebon et al. 2014).
Create new templates
It is possible to create a new template from the anatomy of a given subject. To do so, right-click on Subject > Use template > Create new template.
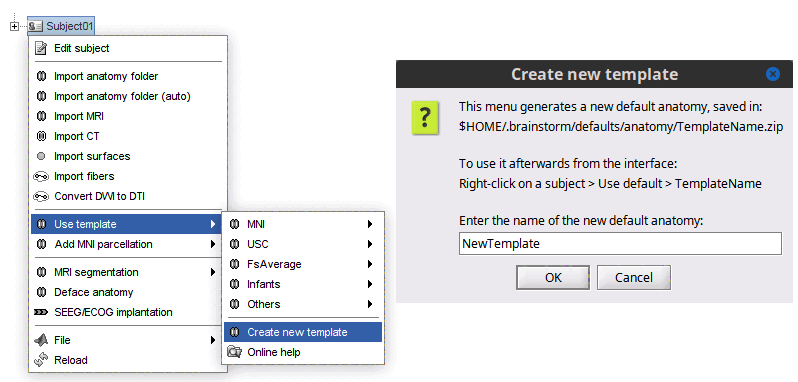
This new template will be saved in $HOME/.brainstorm/defaults/anatomy. Thus it is available in all protocols in Brainstorm. The new template will be located in these groups, depending on it name:
MNI: Name contains icbm or colin
USC: Name contains usc or bci-dni
FsAverage: Name contains fsaverage
Infants: Name contains oreilly or kabdebon or infant
Others: None of the above
MNI parcellations
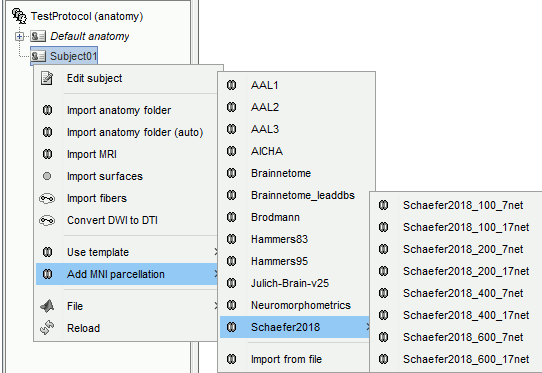
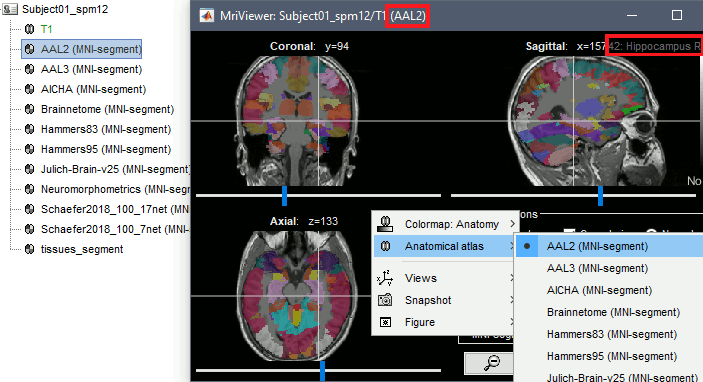
It is possible to import anatomical parcellations of the brain defined in MNI space into any subject anatomy for which the MNI normalization (linear or non-linear) was computed, or into any MNI anatomy template. Each parcellation is a volume of integers where each value represents an anatomical label, and registered to an MNI space. For a subject anatomy which MNI normalization was computed, the MNI parcellation in the subject space is obtained by reslicing the MNI parcellation into the subject space. This can be done for subjects with either linear or non-linear MNI normalization. The anatomical regions defined in these files can be used as volume scouts, for reference in the MRI viewer, or for labelling SEEG contacts.
Right-click on the anatomy folder > Add MNI parcellation. There are some MNI parcellations available for download directly from the Brainstorm interface:
AAL: Website
AICHA: Website
Brainnetome: Two quite versions are available, one coming from the Brainnetome website, and one found in the Lead-DBS distribution.
Brodmann: Brodmann areas, distributed as part of the MRIcron software.
Hammers: Website
Julich-Brain v2.5: Website
Neuromorphometrics: Website
Schaefer2018: Website
You can find detailed descriptions of these MNI parcellations, and link to many other atlases here: