Tutorial 22: Source estimation [TODO]
Authors: Francois Tadel, Elizabeth Bock, Rey R Ramirez, John C Mosher, Richard Leahy, Sylvain Baillet
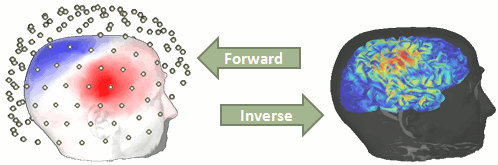
You have in your database a forward model that explains how the cortical sources determine the values on the sensors. This is useful for simulations, but what we need is to build the inverse information: how to estimate the sources when we have the recordings. This tutorials introduces the tools available in Brainstorm for solving this inverse problem.
WARNING: The methods presented here use some new functions that are not fully tested (menu "Compute sources 2016"). For more stable results, it is advised that you keep on using the previous version of these functions (menu "Compute sources", described in the ?old tutorials).
Contents
- Ill-posed problem
- Source estimation options [TODO]
- Computing sources for a single average
- Display: Cortex surface
- Display: MRI Viewer
- Display: MRI 3D
- Sign of constrained minimum norm values
- Unconstrained dipoles orientation
- Computing sources for multiple trials
- Source map normalization [TODO]
- Averaging in source space
- Averaging normalized maps
- Display: Contact sheets and movies
- Advanced options [TODO]
- Model evaluation [TODO]
- Equations [TODO]
- On the hard drive
- References [TODO]
- Additional documentation
Ill-posed problem
Our goal is to estimate the activity of the thousands of dipoles described by our forward model. However we only have a few hundred variables in input (the number of sensors). This inverse problem is ill-posed, meaning there is an infinite number of combinations of source activity patterns that can generate exactly the same sensor topography. Inverting the forward model directly is impossible, unless we add some strong priors to our model.
Wikipedia says: "Inverse problems are some of the most important and well-studied mathematical problems in science and mathematics because they tell us about parameters that we cannot directly observe. They have wide application in optics, radar, acoustics, communication theory, signal processing, medical imaging, computer vision, geophysics, oceanography, astronomy, remote sensing, natural language processing, machine learning, nondestructive testing, and many other fields."
Many solutions have been proposed in the literature, based on different assumptions on the way the brain works and depending on the amount of information we already have on the effects we are studying. Among the hundreds of methods available, two classes of inverse models have been widely used in MEG/EEG source imaging in the past years: minimum-norm solutions and beamformers.
Both approaches have the advantage of being linear: the activity of the sources is a linear recombination of the MEG/EEG recordings. It is possible to solve the inverse problem independently of the recordings, making the data manipulation a lot easier and faster.
Both are available in Brainstorm, so you can use the one that is most adapted to your recordings or to your own personal expertise. Only the minimum norm estimates will be described in this tutorial, but the other solutions work exactly the same way.
Source estimation options [TODO]
Before we start estimating the sources for the recordings available in our database, let's start with an overview of the options available. The screen capture below represents the basic options for the minimum norm estimates. The options for the other methods will be described in advanced tutorials.
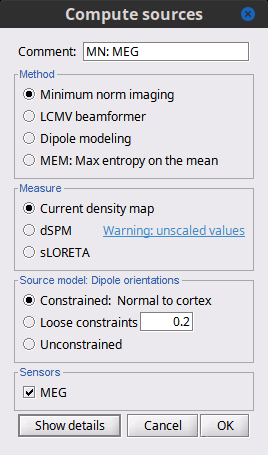
Method [TODO]
Minimum norm: Priors, justification, application case?
Require an estimation of the noise at the level of the sensors (noise covariance matrix).LCMV beamformer: ?
Require both a noise covariance matrix and a data covariance matrix (representation of the effect we are trying to localize in the brain, covariance of the latencies of interest).Dipole modeling: ?
Warning: This is not a imaging method!Recommended option: Provided that we know at which latencies to look at, we can compute a correct data covariance matrix and may obtain a better spatial accuracy with a beamformer. However, in many cases we don't exactly know what we are looking at, therefore the risks of misinterpretation of badly designed beamforming results are high. Brainstorm tends to favor minimum norm solutions, which have the advantage of needing less manual tuning for getting acceptable results.
Measure [TODO]
The minimum norm estimates produce a measure of the current density flowing at the surface of the cortex. To visualize these results and compare them between subjects, we can normalize the MNE values to get a standardized level of activation with respect to the noise or baseline level (dSPM, sLORETA, etc).
Current density map: Whitened and depth-weigthed linear L2-minimum norm estimates algorithm inspired from Matti Hamalainen's MNE software. For a full description of this method, please refer to the MNE manual, section 6, "The current estimates".
Units: picoamper per meter (pA.m).dSPM: Noise-normalized estimate (dynamical Statistical Parametric Mapping [Dale, 2000]). Its computation is based on the MNE solution.
Units: Unitless ratio [ ??? ]sLORETA: Noise-normalized estimate using the sLORETA approach (standardized LOw Resolution brain Electromagnetic TomogrAphy [Pasqual-Marqui, 2002]). sLORETA solutions have, in general, a smaller location bias than either the expected current (MNE) or the dSPM. The noise covariance is not used at all for the standardization process, it is purely based on the smoothness of the maps.
Units: Square root of units of current (MNE/sqrt(MNE) => (pA.m)1/2).Recommended option: Discussed in the section "Source map normalization" below.
Source orientation
Constrained: Normal to cortex: Only one dipole at each vertex of the cortex surface, oriented normally to the surface. This is based on the anatomical observation that in the cortex, the neurons are mainly organized in macro-columns that are perpendicular to the cortex surface.
Size of the inverse operator: [Nvertices x Nchannels].Unconstrained: At each vertex of the cortex surface, we define a base of three dipoles with orthogonal directions, then we estimate the sources for the three orientations independently.
Size of the inverse operator: [3*Nvertices x Nchannels].Loose: A version of the "unconstrained" option with a weak orientation constraint that emphasizes the importance of the sources with orientations that are close to the normal to the cortex. The value associated with this option set how "loose" should be the orientation constrain (recommended values in MNE are between 0.1 and 0.6, --loose option).
Size of the inverse operator: [3*Nvertices x Nchannel].Recommended option: The constrained options use one dipole per grid point instead of three, therefore the source files are smaller, faster to compute and display, and more intuitive to process because we don't have to think about recombining the three values into one. On the other hand, in the cases where its physiological assumptions are not verified, the normal orientation constraint may fail representing certain activity patterns. Unconstrained models can help in those cases.
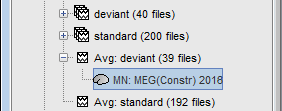
Computing sources for a single average
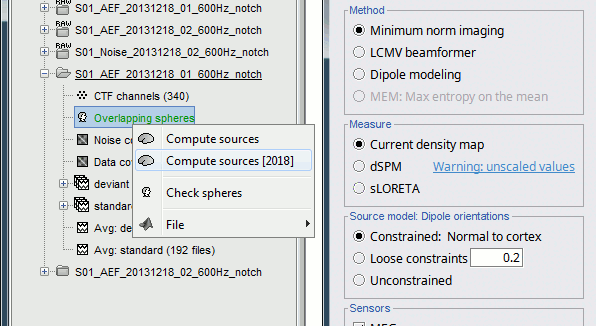
In Run#01, right-click on the average response for the deviant stim > Compute sources [2016].
Select the options: Minimum norm imaging, Current density map, Constrained: Normal to cortex.
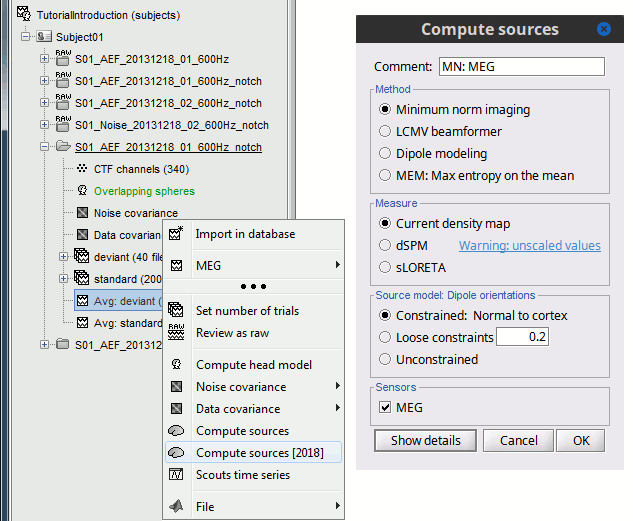
- The other menu "Compute sources" brings the interface that was used previously in Brainstorm. We are going to keep maintaining the two implementations in parallel for a while for compatibility and cross-validation purposes.
The result of the computation is displayed as a dependent file of the deviant average because it is related only to this file. In the file comment, "MN" stands for minimum norm and "Constr" stands for "Constrained: normal orientation".
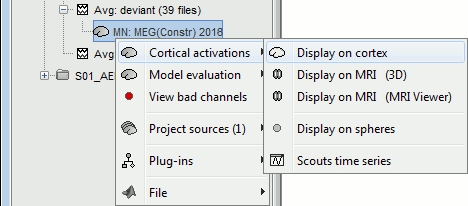
Display: Cortex surface
Right-click on the sources for the deviant average > Cortical activations > Display on cortex.
Double-click on the recordings for the deviant average to have time reference.
In the filter tab, add a low-pass filter at 40Hz.
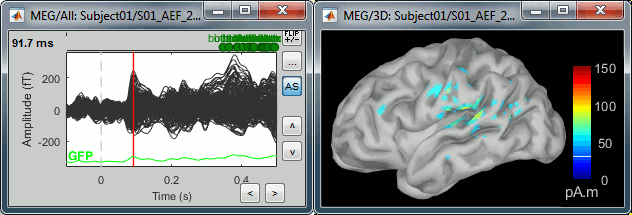
- Change the current time (click on the time series figure or use the keyboard arrows) and note it updates the source maps in the 3D figure. You can also use all the menus and shortcuts introduced in the anatomy tutorial (like setting the view with the keys from 0 to 6).
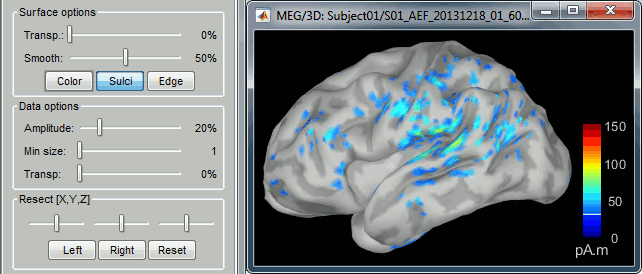
- You can edit the display properties in the Surface tab:
Amplitude: Only the sources that have a value superior to a given percentage of the colorbar maximum are displayed.
Min. size: Hide all the small activated regions, ie. the connected color patches that contain a number of vertices smaller than this "min size" value.
Transparency: Change the transparency of the source activity on the cortex surface.
Take a few minutes to understand what the amplitude threshold represents.
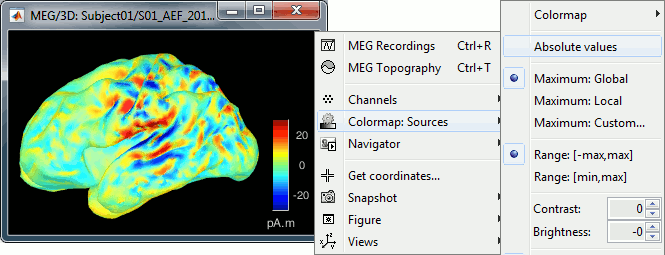
The colorbar maximum depends on the way you configured your Sources colormap. If the option "Maximum: Global" is selected, the maximum should be around 130 pA.m. This value is a rough estimate of the maximum amplitude, and this default value is not always adapted to your figure. To edit the maximum value, use the colormap option "Maximum: Custom".
On the screen capture below, the threshold value is set to 20%. It means that only the sources that have a value over 0.20*130 = 26 pA.m are visible.
The threshold level is indicated in the colorbar with a horizontal white line.
At the first response peak (91ms), the sources with high amplitudes are located around the primary auditory cortex, bilaterally, which is what we are expecting for an auditory stimulation.
Display: MRI Viewer
Right-click on the source file > Cortical activations > Display on MRI (MRI Viewer).
The MRI viewer was introduced in tutorials #2 and #3.
Additionally you can change the current time and amplitude threshold from the Brainstorm window.This figure shows the sources computed on the cortical surface and re-interpolated in the MRI volume. If you set the amplitude threshold to 0%, you would see the thin layer of cortex in which the dipoles where estimated.
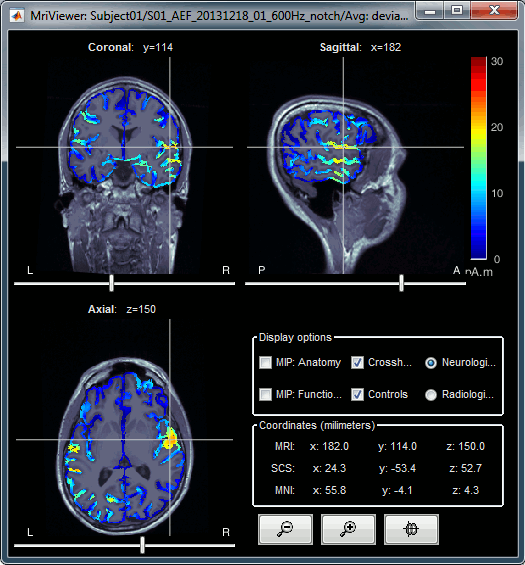
- In most of the screen captures in this section and the following, the contrast of the figures has been enhanced for illustration purposes. Don't worry if it looks a lot less colorful on your screen.
- You can configure this figure with the following options:
MIP Anatomy: Checkbox in the MRI Viewer figure. For each slice, display the maximum value over all the slices instead of the original value in the structural MRI ("glass brain" view).
MIP Functional: Same as for MIP Anatomy, but with the layer of functional values.
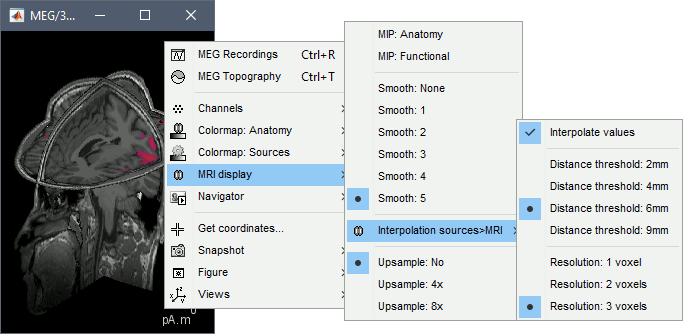
Smooth level: The sources values can be smoothed after being re-interpolated in the volume. Right-click on the figure to define the size of the smoothing kernel.
Amplitude threshold: In the Surface tab of the Brainstorm window.
Current time: At the top-right of the Brainstorm window (or use the time series figure).
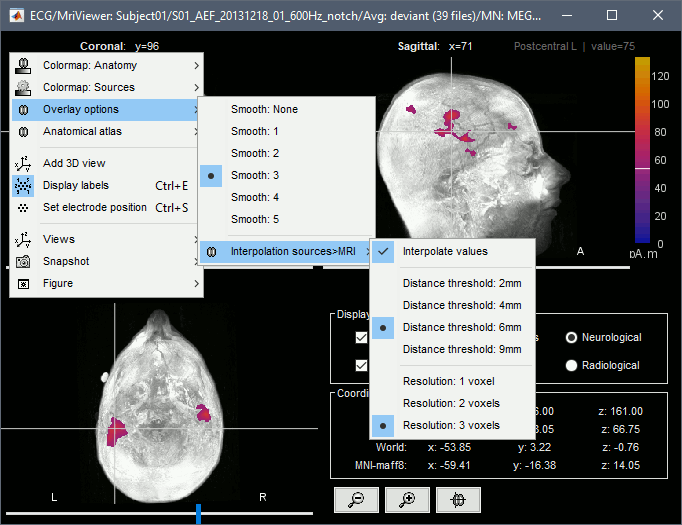
Display: MRI 3D
Right-click on the source file > Cortical activations > Display on MRI (3D).
This view was also introduced in the tutorials about MRI and surface visualization.
Right-click and move your mouse to move the slices (or use the Resect panel of the Surface tab).
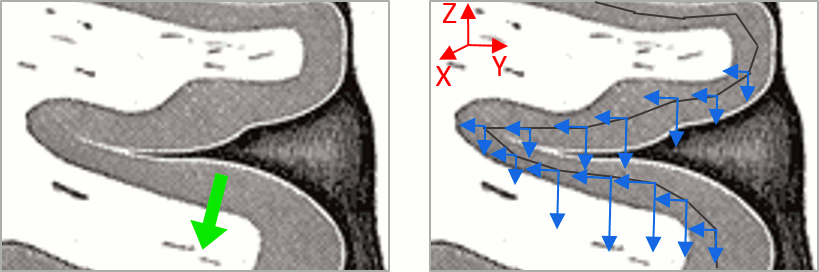
Sign of constrained minimum norm values
You should pay attention to the sign of the current amplitudes that are given by the minimum norm method: they can be positive or negative and they oscillate around zero. Display the sources on the surface, set the amplitude threshold to 0%, then configure the colormap to show relative values (uncheck the "Absolute values" option), you would see those typical stripes of positive and negative values around the sulci. Double-click on the colorbar after testing this to reset the colormap.
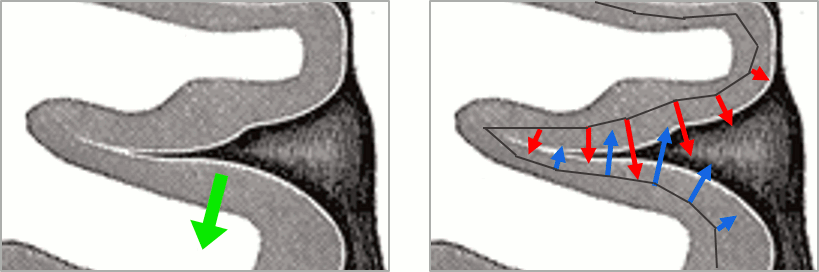
This pattern is due to the orientation constraint imposed on the dipoles. On both sides of a sulcus, we have defined dipoles that are very close to each other, but with opposite orientations. If we have a pattern of activity on one side of a suclus that can be assimilated to an electric dipole (green arrow), the minimum norm model will try to explain it with the dipoles that are available in the head model (red and blue arrows). Because of the dipoles orientations, it translates into positive values (red arrows) on one side of the sulcus and negative on the other side (blue arrows).
When displaying the cortical maps at one time point, we are usually not interested by the sign of the minimum norm values but rather by their amplitude. This is why we always display them by default with the colormap option "absolute values" selected.
However, we cannot simply discard the sign of these values because we need them for other types of analysis, typically time-frequency decompositions and connectivity analysis. For estimating frequency measures on the source maps, we need to keep the oscillations around zero.
Unconstrained dipoles orientation
Double-click on the new link for the deviant average, to see what unconstrained source maps look like. The first obvious observation is that the maps look a lot smoother.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
We have to be careful with the visual comparisons of constrained and unconstrained source maps displayed on the cortex surface, because they are very different types of data. In unconstrained source maps, we have three dipoles with orthogonal orientations at each cortex location, therefore we cannot represent all the information at once. To display them as an activity map, Brainstorm computes the norm of the vectorial sum of the three orientations at each vertex.
Same for "loose"
Computing sources for multiple trials
Because the minimum norm model is linear, we can compute an inverse model independently from the recordings and apply it on the recordings when needed. We will now illustrate how to compute a shared inverse model for all the imported epochs. For illustration purpose, we will use this time an unconstrained source model.
Right-click on the head model or the folder for Run#01 > Compute sources [2016].
Select the options: Minimum norm imaging, Current density map, Unconstrained
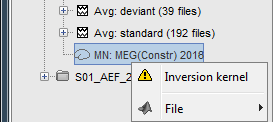
Because we did not request to compute and inverse model for a specific block of recordings, it computed a shared inverse model. If you right-click on this new file, you get a warning message: "Inversion kernel". It does not contain any source map, but only the inverse operator that will allow us to convert the recordings in source maps.
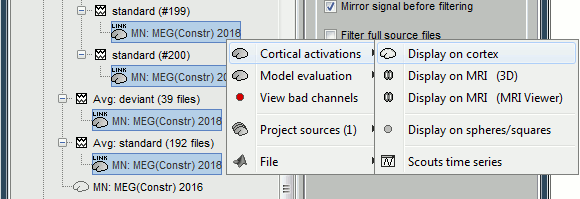
The database explorer now shows one source link to this inverse model for each block of recordings available in this folder, single trials and averages. These links are not real files saved on the hard drive, but you can use them exactly like the first source file we calculated for the deviant average. If you load a link, Brainstorm loads the corresponding MEG recordings, loads the inverse kernel and multiplies the two on the fly before displaying it. This optimized approach saves a lot of computation time and lot of space on the hard drive.
Source map normalization [TODO]
The current density values returned by the minimum norm method have a few problems:
They depend a lot on the SNR of the signal, which may vary significantly between subjects.
Their amplitude is therefore difficult to interpret directly.- The values tend to be always higher at the surface of the brain (close to the sensors).
- The maps are sometimes patchy and difficult to read.
Normalizing the current density maps with respect to a baseline (noise recordings or resting state) can help with all these issues at the same time. Some normalizations can be computed independently from the recordings, and saved as part of the linear source model (dSPM, sLORETA, MNp). Another way of proceeding is to compute a Z-score baseline correction from the current density maps.
The normalization options do not change your results, they are just different ways for looking at the same minimum norm maps. If you look at the time series associated with one source, it would be exactly the same for all the normalizations, except for a scaling factor. Only the relative weights change between the sources, and these weights do not change over time.
dSPM, sLORETA, MNp
In Run#01, right-click on the average recordings for the deviant stim > Compute sources [2015].
Select successively the three normalization options: dSPM, sLORETA, MNp (unconstrained).
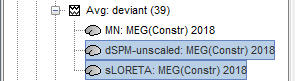
Double-click on all of them to compare them:
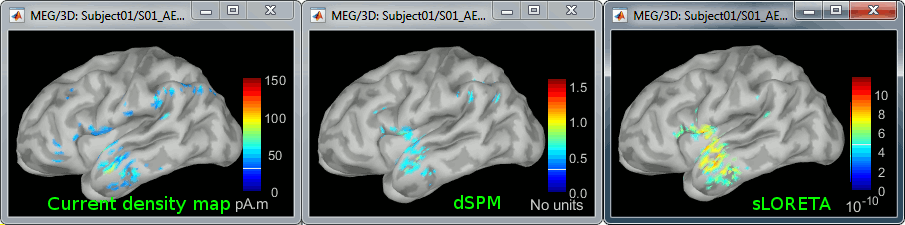
Current density maps: Tends to highlight the top of the gyri and the superficial sources.
dSPM: Tends to correct that behavior and may give higher values in deeper areas.
sLORETA: produces a very smooth solution where all the potentially activated area of the brain (given to the low spatial resolution of the source localization with MEG/EEG) is shown as connected, regardless of the depth of the sources.
MNp: ???
Z-score
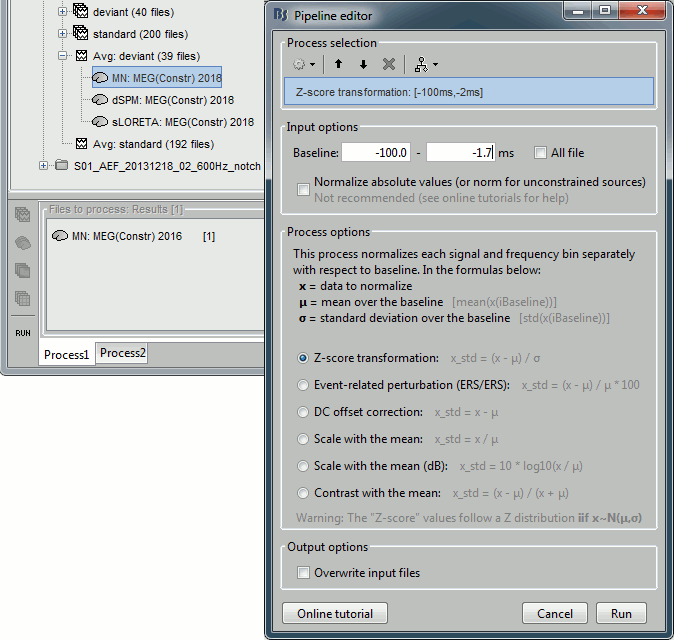
The Z-transformation converts the current density values to a score of deviation from a baseline. For each source separately we define a baseline, compute the average and standard deviation for this segment. Then for every time point we subtract the baseline average and divide by the baseline standard deviation. It tells how much a value deviates from the average at rest (in number of times the standard deviation).
- Drag and drop the unconstrained current density maps (the only link) to the Process1 list.
Run process "Standardize > Z-score normalization", baseline = [-100,-2]ms.
Select the option "Use the norm of the three orientations", if not it would compute the Z-score normalization separately for each direction and then take the norm of the three Z-scored orientation for display, which doesn't make much sense.
The option "dynamic" offers an optimization in the storage of the Z-scored file that can save a lot of disk space in some cases, but that is incompatible with the "norm" option for unconstrained sources.
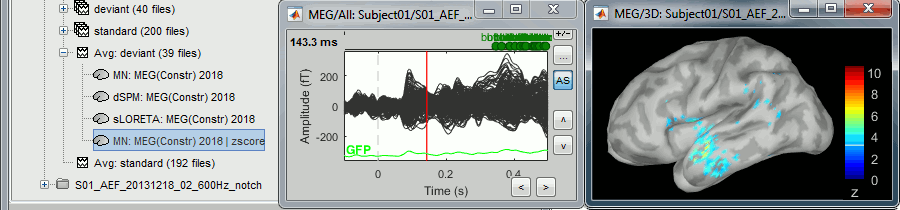
The option "absolute values / norm of the three orientations" caused the call to the intermediate process "Sources > Unconstrained to flat map: norm". This is why the comment of the output file includes the tag "norm".
Note that this process brought down the number of signals in the file from 45,000 (the number of dipoles) to 15,000 (the number of grid points). There is now only one normalized value for each vertex of the cortex surface.
If the baseline and the active state are not in the same file, you can use the Process2 tab: place the baseline in the left list (Files A) and the file to normalize in the right list (Files B).
Delete your experiments
Select all the normalized source maps (everything but the link) and delete them.
Typical recommendations
- Use non-normalized current density maps for:
- Computing shared kernels applied to single trials.
- Averaging single trials across MEG runs.
- Computing time-frequency decompositions or connectivity measures on the single trials.
- Use normalized maps (dSPM, sLORETA, MNp, Z-score) for:
- Estimating the sources for an average response.
- Exploring visually the ERP/ERF at the source level.
- Normalizing the subjects averages before a group analysis.
Averaging in source space
Computing the average
First compute the same source model for the the second acquisition run.
In Run#02, right-click on the head model or the folder > Compute sources [2015].
Select the options: Minimum norm imaging, Current density map, Unconstrained
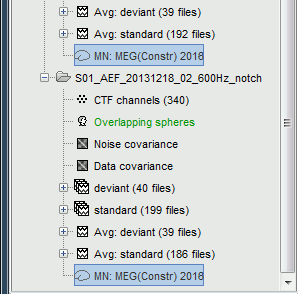
Now we have the source maps available for all the trials, we can average them in source space across runs. This allows us to average MEG recordings that were recorded with different head positions (in this case Run#01 and Run#02 have different channel files so they could potentially have different head positions preventing the direct averaging at the sensor level).
- Thanks to the linearity of the minimum norm model, the two following approaches are equivalent:
- 1. Averaging the sources of all the individual trials across runs,
- 2. Averaging the sources for the sensor averages that we already computed for each run. For non-standardized current density maps: MN(Average(trials)) = Average(MN(trials))
- The second solution is a lot faster because it needs to read 4 files (one file per run and per condition) instead of 39*4 = 156 (we used 39 trials in each condition and each run).
Number of trials: Because we want to compare directly the two conditions deviant and standard, we should use the same number of trials in both averages. If you were computing the source average directly from the single trials, you could use the process "File > Select uniform number of trials", as we did when computing the sensor average (see Tutorial 16: Average response).
Drag and drop to the Process1 tab the average recordings for Run01 and Run02, then press the [Process sources] button on the left.
Run process "Average > Average files": Select "By trial group (subject average)".
The options "trial group" average together the files that have similar comments.
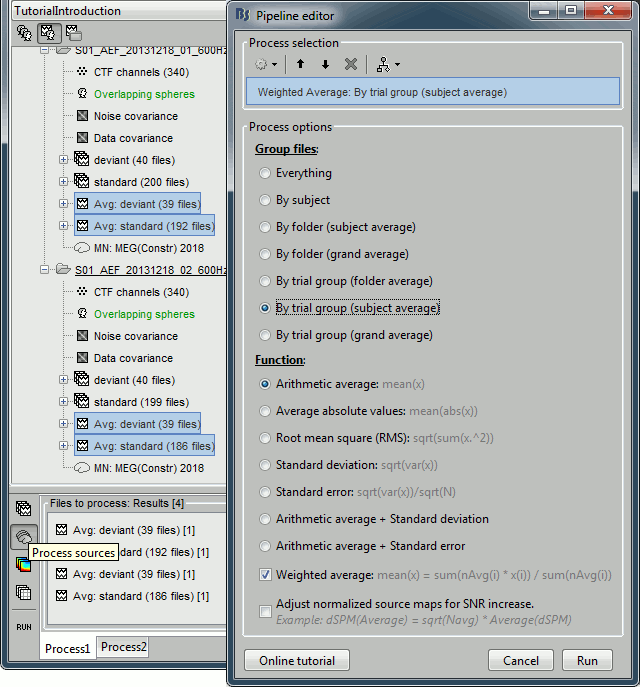
The two averages that are produced (one for each condition) are saved in the folder Intra-subject. This is where all the files that were computed using information from multiple folders within the same subject are going to be saved. If you prefer to have them in different folders, you can create new folders and move them there.
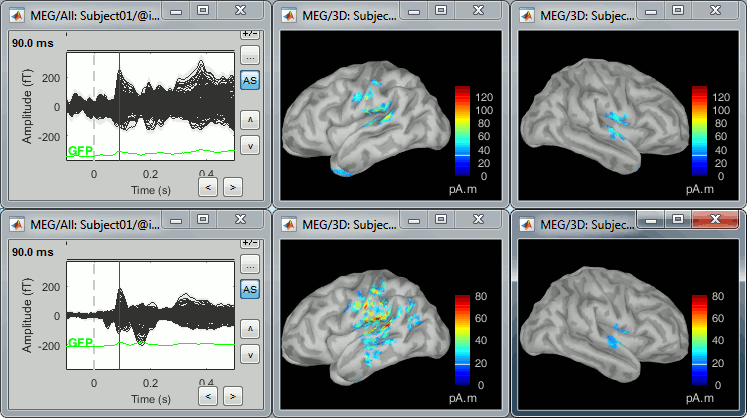
Double-click on the source averages to display them (standard=top, deviant=bottom).
Open the average recordings of one of the runs as a time reference.
Use the pre-defined view "Left, Right" for looking at the two sides at once (shortcut: "7").
Low-pass filter
Note that opening the source maps can be very long because of the filters for visualization. Check in the Filter tab, you may have a filter applied with the option "Filter full source files" selected. In the case of averaged source maps, the 45,000 source signals are filtered on the fly when you load a source file. This filtering of the full source files can take a significant amount of time, consider unchecking this option if the display is too slow on your computer.
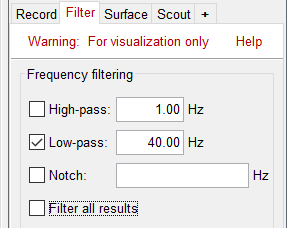
- It was not a problem until now because the source files were saved in the compact form (Kernel*recordings) and the visualization filters were applied on the recordings, then projected to the source space.
- These visualization filters will not be available anymore after we apply a Z-score normalization. If we want to display source maps that are smoothed in time, this is the last moment were we can apply a low-pass filter.
Clear the Process1 list, then drag and drop the new averages in it.
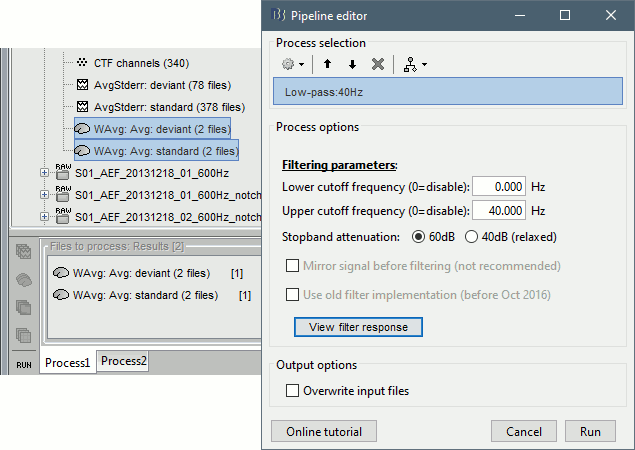
Select process "Pre-process > Band-pass filter": [0,40] Hz
Do not run the computation, we will add the Z-score process immediately after.
Z-score normalization
Add process "Standardize > Z-score normalization", baseline = [-75,-2]ms.
We do not consider the entire 100ms baseline because of the low-pass filter: it may have created edge effects during the first and last 1/40=25ms.
Select the option "Use the norm of the three orientations"
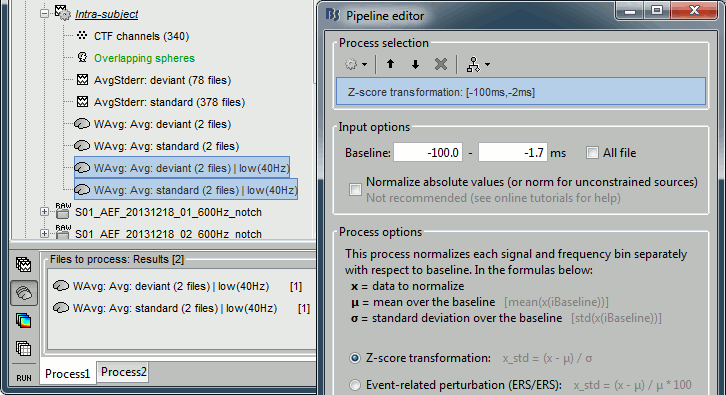
Four new files are accessible in the database: two filtered and two filtered+normalized.
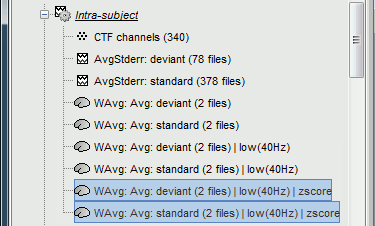
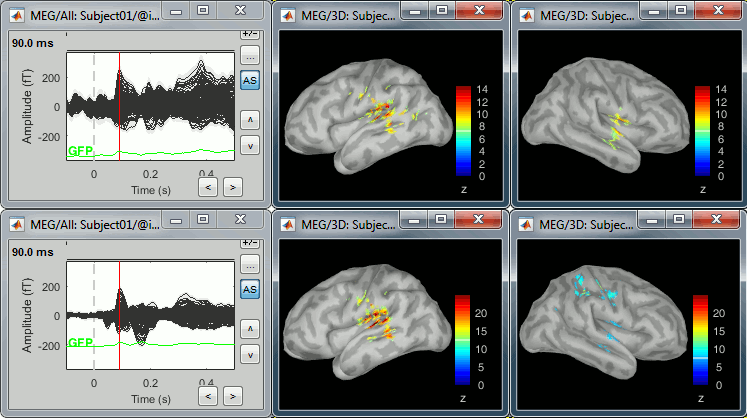
Double-click on the source averages to display them (standard=top, deviant=bottom).
Averaging normalized maps
Averaging normalized values such as dSPM source maps requires more attention than averaging current density maps. The amplitude of the measures increase with the SNR of the signal, the higher the SNR the higher the dSPM score. The average of the dSPM score for the single trials is lower than the dSPM of the averaged trials.
MinNorm(Average(trials)) = Average(MinNorm(trials))
dSPM(Average(trials)) = sqrt(Ntrials) * Average(dSPM(trials))
When computing the average of dSPM or other normalized values, we have to also multiply the average with the square root of the number of files averaged together. To illustrate this, we can compute dSPM values of the averages for each run, and then average the two runs together.
Drag and drop to the Process1 tab the average recordings for Run01 and Run02, then press the [Process recordings] button on the left.
Select process "Sources > Compute sources [2015]"
Select the option "Kernel only: One per file", then click on the [Edit] button.
Select Method=Minimum norm, Measure=dSPM, Source model=Unconstrained.
Do not run the process immediately.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
Add process "Average > Average files": Select "By trial group (subject average)".
Select the option "Adjust normalized source maps for SNR increase".
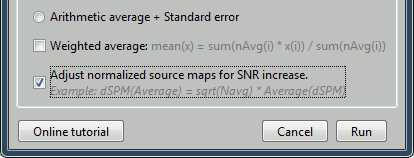
The option weighted average should be selected in general when re-averaging run-level averages. However, in this case we selected the same number of trials for all the conditions, so the regular and weighted averages are the same.
At the end, the report viewer shows the warning: "Averaging normalized maps (dspm): scaling the values by sqrt(78/39)=sqrt(2)=1.414 to match the number of trials averaged (39 => 78)".
Nothing went wrong, this is just to make it clear that the final averaged values have been scaled.Rename the two last files to remember that they correspond to dSPM averages, not current maps.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
Double-click on the files to display them. You can note that the cortical maps for the primary response are very similar for the two conditions (left=deviant, right=standard), which matches our hypothesis because at this early stage we should not observe any significant difference.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
For a comparison, this is what we would get if we were averaging the source without selecting the option "Adjust normalized source maps for SNR increase". Exactly the same maps but with lower amplitudes: same values without the 1.414 factor that was mentioned in the warning.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
Delete all the dSPM files you've just computed, in the two runs and in Intra-subject, to avoid confusion between different source models in the next tutorials.
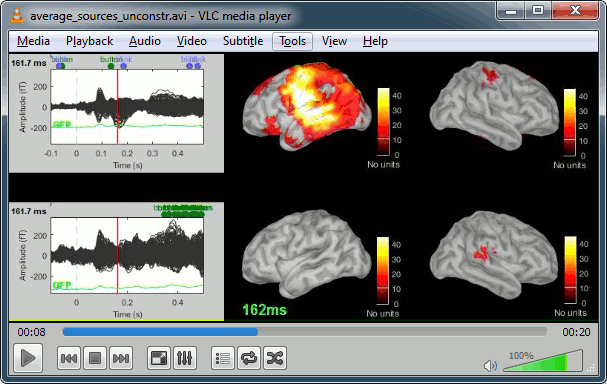
Display: Contact sheets and movies
A good way to represent what is happening in time is to generate contact sheets or videos. Right-click on any figure and go to the menu Snapshot to check out all the possible options. For a nicer result, take some time to adjust the size of the figure, the amplitude threshold and the colormap options (hiding the colorbar can be a good option for contact sheets).
A time stamp is added to the captured figure. The size of the text font is fixed, so if you want it to be readable in the contact sheet, you should make you figure very small before starting the capture.
Contact sheet: Right-click on any figure > Snapshot > Time contact sheet: Figure

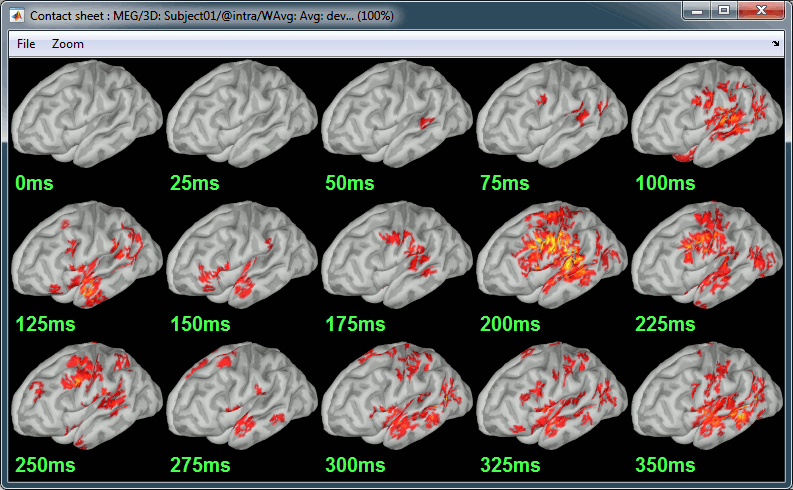
The propagation of the deviant detection from temporal to frontal, in more details:
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
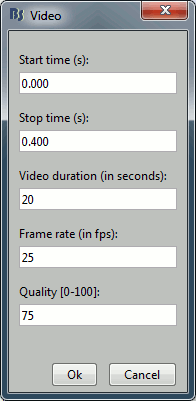
Movies: Right-click on any figure > Snapshot > Movie (time): All figures (click to download video)
Advanced options [TODO]
Right-click on the deviant average for Run#01 > Compute sources [2015].
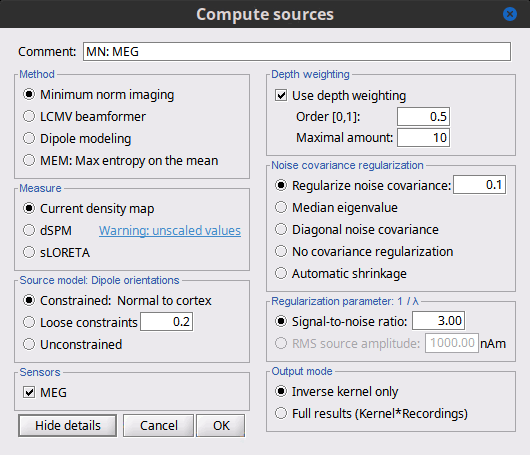
Click on the button [Show details] to bring up all the advanced minimum norm options.
Depth weighting
The minimum-norm estimates have a bias towards superficial currents. This tendency can be alleviated by adjusting these parameters. To understand how to set these parameters, please refer to the MNE manual (options --depth, --weightexp and --weightlimit).
Noise covariance regularization
Explain the influence of this parameter.
Automatic shrinkage:
Regularize noise covariance: Regularize the noise-covariance matrix by the given amount for each type of sensor individually (value is restricted to the range 0...1). For more information, please refer to the MNE manual, section 6.2.4 (options --magreg, --gradreg and --eegreg).
Diagonal noise covariance:
None:
Signal-to-noise ratio
Explain the influence of this parameter.
RMS source amplitude:
Use fixed SNR:
Estimate SNR from data:
Output mode
Full results: Saves in one big matrix the values of all the sources (45,000) for all the time samples (361). The size in memory of such a matrix is about 130Mb for 600ms of recordings. This is still reasonable, so you may use this option in this case. But if you need to process longer recordings, you may face "Out of memory" errors in Matlab, or fill your hard drive quickly.
Kernel only: Saves only the linear inverse operator, a model that converts sensor values into source values. The size of this matrix is: number of sources (45000) x number of MEG sensors (274). The multiplication with the recordings is done on the fly by Brainstorm in a transparent way. For long recordings or numerous epochs, this form of compact storage helps saving a lot of disk space and computation time, and it speeds up significantly the display. Always select this option when possible.
- Full results [45000x361] = Inverse kernel [45000x274] * Recordings [274x361]
Dipole modeling
Explain the method briefly.
Best dipole fit:
Goodness-of-fit map:
Chi-square error map:
NP performance index:
LCMV beamformer
Explain the method briefly.
Beamformer time-series:
Beamformer power:
Neural activity index:
NP performance index:
Model evaluation [TODO]
...
Equations [TODO]
...
On the hard drive
Unconstrained shared kernel
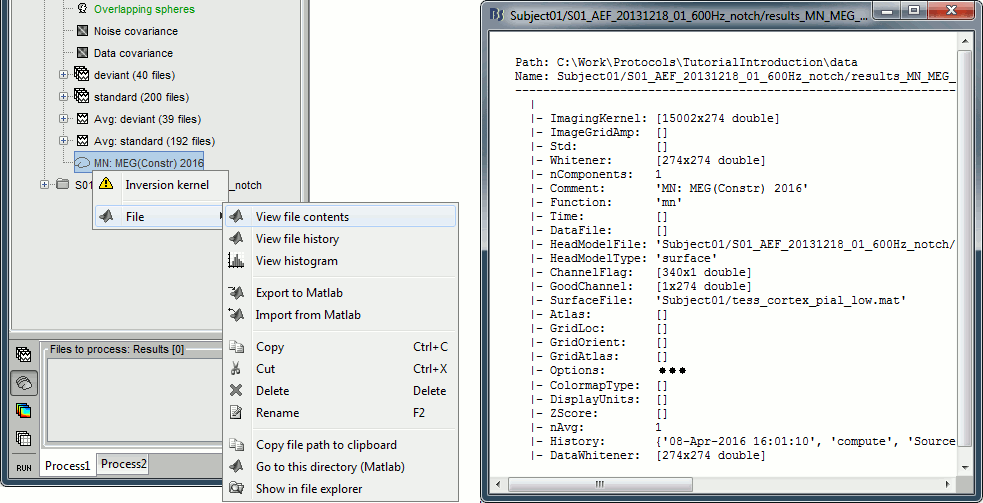
Right-click on a shared inverse file in the database explorer > File > View file contents.
Structure of the source files: results_*.mat
Mandatory fields:
ImagingKernel: [Nsources x Nchannels] Linear inverse operator that must be multiplied by the recordings in order to get the full source time series. If defined, ImageGridAmp must be empty.
ImageGridAmp: [Nsources x Ntime] Full source time series, in Amper.meter. If this field is defined, ImagingKernel must be empty.
Time: [1 x Ntime] Time values for each sample recorded in F, in seconds.
nComponents: Number of dipoles per grid point: 1=Constrained, 3=Unconstrained, 0=Mixed. In the case of mixed head models, the atlas GridAtlas documents region by region how the list of grid points matches the list of dipoles.
Function: Type of values currently saved in the file: 'mn', 'mnp', 'dspm', 'sloreta', 'lcmv', 'lcmvp', 'lcmvnai', 'lcmvpow', 'gls', 'glsp', 'glsfit', 'glschi', 'zscore', 'ersd'...
HeadModelType: Type of source space used for this head model ('surface', 'volume', 'mixed').
HeadModelFile: Relative path to the head model used to compute the sources.
SurfaceFile: Relative path to the cortex surface file related with this head model.
Atlas: Used only by the process "Sources > Downsample to atlas".
GridLoc: [Nvertices x 3], (x,y,z) positions of the grid of source points. In the case of a surface head model, it is empty and you read directly the positions from the surface file.
GridOrient: [Nvertices x 3], direction of the normal to the surface for each vertex point (copy of the 'VertNormals' matrix of the cortex surface). Empty in the case of a volume head model or unconstrained sources.
GridAtlas: Atlas "Source model" used with mixed source models.
GoodChannel: [1 x Nchannels] Array of channel indices used to estimate the sources.
DataFile: Relative path to the recordings file for which the sources where computed. If this field is set, the source file appears as a dependent of the DataFile.
Comment: String displayed in the database explorer to represent this file.
History: Operations performed on the file since it was create (menu "View file history").
Optional fields:
Options: Structure that contains all the options of the inverse calculation. This is saved in the file only for bookkeeping.
Whitener: Noise covariance whitener computed in bst_inverse_linear_2015.m
DataWhitener: Data covariance whitener computed in bst_inverse_linear_2015.m
Std: For averaged files, number of trials that were used to compute this file.
DisplayUnits: String, force the display of this file using a specific type of units.
ChannelFlag: [Nchannels x 1] Copy of the ChannelFlag field from the original data file.
nAvg: For averaged files, number of trials that were used to compute this file. For source files that are attached to a data file, we use the nAvg field from the data file.
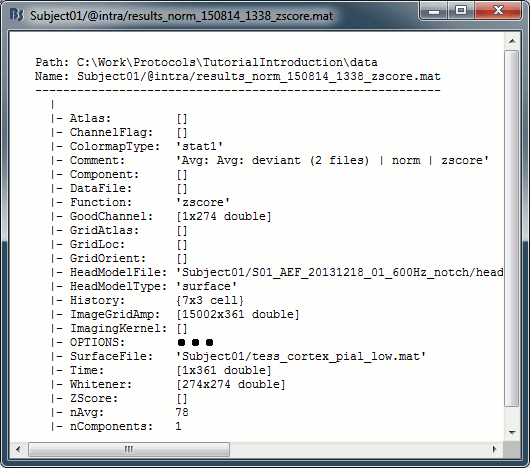
Flattened full source maps
In Intra-subject, right-click on one of the normalized averages > File > View file contents.
This file has the same structure as a shared inverse kernel, with the following differences:
This file contains the full time series (ImageGridAmp) instead of an inverse operator (ImagingKernel).
The Z-score process computed the norm of the three unconstrained orientations, so ImageGridAmp only describes 15,000 signals instead of 45,000 previously.
The Z-score process updated the field Function ('mn' => 'zscore')
Source links
- The links are not real files on the hard drive, if you select the menu "View file contents" for any of them it would display the structure of the corresponding shared kernel.
They are saved in the database as strings with a specific structure: "link|kernel_file|data_file". This string associates a shared inverse operator with some recordings. The two files have to be available to load the this file. All the functions in Brainstorm are equipped to reconstruct the full source matrix dynamically.
Filename tags
_KERNEL_: Indicates that the file contains only an inverse kernel, it needs to be associated with recordings to be opened.
Useful functions
in_bst_results(ResultsFile, LoadFull, FieldsList): Load a source file and optionnally reconstruct the full source time series on the fly (ImagingKernel * recordings).
in_bst(FileName, TimeBounds, LoadFull): Load any Brainstorm data file with the possibility to load only a specific part of the file.
bst_process('LoadInputFile', FileName, Target, TimeWindow, OPTIONS): The most high-level function for reading data files, can compute scout values on the fly.
References [TODO]
Dale AM, Liu AK, Fischl BR, Buckner RL, Belliveau JW, Lewine JD, Halgren E
Dynamic statistical parametric mapping: combining fMRI and MEG for high-resolution imaging of cortical activity. Neuron 2000 Apr, 26(1):55-67Pascual-Marqui RD, Standardized low-resolution brain electromagnetic tomography (sLORETA): technical details, Methods Find Exp Clin Pharmacol 2002, 24 Suppl D:5-12
Additional documentation
Tutorial: Volume source estimation
Tutorial: Deep cerebral structures
Tutorial: Computing and displaying dipoles
Tutorial: Dipole fitting with FieldTrip
Tutorial: Maximum Entropy on the Mean (MEM)
Forum: Minimum norm units (pA.m): http://neuroimage.usc.edu/forums/showthread.php?1246
Forum: Imaging resolution kernels: http://neuroimage.usc.edu/forums/showthread.php?1298
Forum: Spatial smoothing of sources: http://neuroimage.usc.edu/forums/showthread.php?1409
Forum: Units for dSPM and sLORETA: http://neuroimage.usc.edu/forums/showthread.php?1535
Forum: EEG reference: http://neuroimage.usc.edu/forums/showthread.php?1525#post6718
Forum: Sign of the MNE values: http://neuroimage.usc.edu/forums/showthread.php?1649#post7014
Forum: Combining magneto+gradiometers: http://neuroimage.usc.edu/forums/showthread.php?1900
Forum: Residual ocular artifacts: http://neuroimage.usc.edu/forums/showthread.php?1272