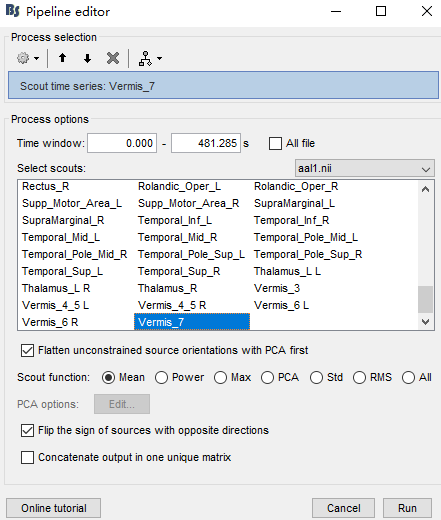
Recently in the use of brainstorm, for source localization related work, after doing the source estimation, I want to extract the time series according to the anatomical template division of ROI, I downloaded the template file of MNIAAL1 from the official website, as shown in the figure, it is reasonable that it should be AAL116, there are a total of 116 ROIs, why do I only show 110 here.Has brainstorm improved this template?
What is the source space for your sources?
(cortical) surface, (brain) volume?
This is correct AAL1 has 116 anatomical parcellations.
The 116 ROIs in AAL116 are defined for voxels in the MNI152 template.
However, once you use that anatomical parcellation to create Brainstorm scouts, which are defined in the source grid rather than in the voxel "grid", the number of Scouts can be smaller:
-
If an anatomical parcellation is only few voxels, it may be the case that there is not correspondence to a vertex in the source grid.
-
If source grid does not include all the space that is included in the AAL1 anatomical parcellation. E.g, AAL1 has ROIs for Thalamus, but your source space is just cortical surface, there will not be source vertices that correspond to Thalamus, thus that Scout will not be present
Continuing the discussion from A question about anatomical templates:
Thank you for your heartfelt answer, I appreciate it. But I still have three questions
Q1:you refer thae if source grid does not include all the space that is included in the AAL1 anatomical parcellation.
Is this related to some brain electrodes I havn't, or am I missing some step?
Q2:So what more processing do I need to do after doing the source estimation to get the appropriate number of ROI time series based on the anatomical template.and get the right number:116
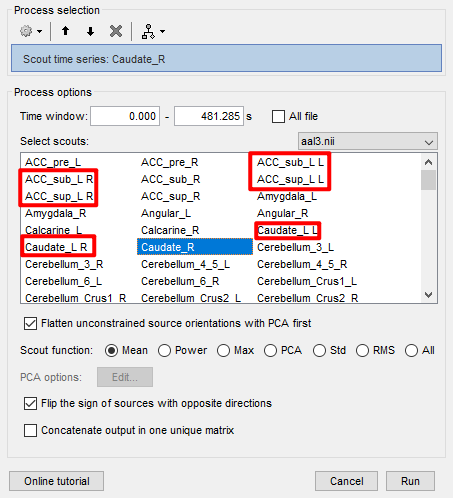
Q3:Why do some of the above template files show errors like ACC_sub_L L ,ACC_sub_L R , ACC_sup_L L, ACC_sup_L R, duplicated display?
What is the source space for your sources?
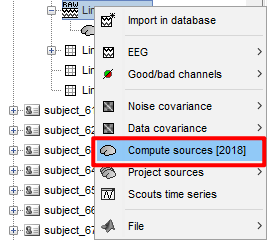
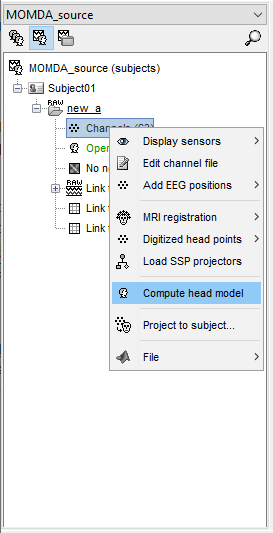
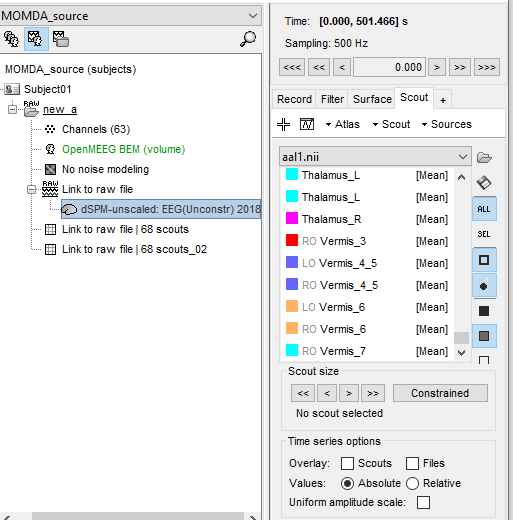
it's (cortical) surface.After this step as shown, I then extracted the time series using the appropriate template
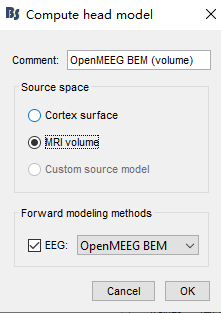
The source space is not set when computing sources, but when you define the head model
Check the section Whole-brain model in
https://neuroimage.usc.edu/brainstorm/Tutorials/HeadModel#Source_models
When you compute the head model, you indicate the source space
- Cortex surface: All source dipoles (points in the source grid) are located on the cortical surface
- MRI volume: Source dipoles are located in a volume grid that is generated, according to the provided parameters. Details in here: https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource#Compute_a_volume_head_model
From your screenshots it seems you are using Cortex surface as source space.
Is this correct?
You would need to make a volume source grid based the inner-skull (or head) with a resolution as high as the MRI volume. In that sense there should be a vertex for each voxel. However, this number of sources is not practical to handle, nor recommended.
First, the shown image is not for AAL1 but for AAL3 which as more ROI
I have the impression that what is going on is that when the ACC_sub_L anatomical parcellation is projected into the cortical surface it contains vertices in the L and R hemispheres, thus two surface scouts are created ACC_sub_L L and ACC_sub_L R.
You can double-check if this is the case, by displaying the surface and showing only those two scouts.
Thanks again for your patience in answering! I would like to check with you again if there are any other issues with the steps I took to extract the corresponding ROIs after performing the EEG source localization, which I did as follows:
- EEG dataset preprocessing
- Create SUBJECT object and load data, load electrode template
- Calculate the head model to choose MRI instead of cortex, in order as follows
4.Noise covariance calculation
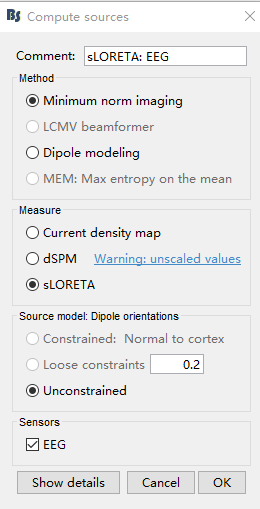
5.Compute Sources
6.According to the anatomical template, such as AAL1, extract the corresponding time series
Because I only have one EEG dataset, I am not sure about other related parameters, so I use default parameters for all source localization related operations.