Dear Brainstormers,
I've been trying to analyze sources files from Brainstorm in SPM,
I exported the files in the "volume" format, and then I did a script in matlab to automatize the exportation process for several conditions and time windows,
I've noted two issues,
-
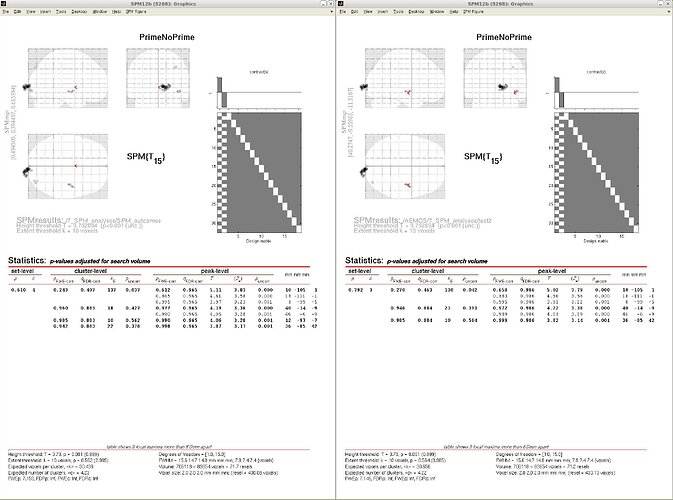
In order to be sure my script works fine, I compared the statistical results when importing manually (with the GUI) and with my script,
In the figure attached (on the left manually imported, on the right imported with the script) you can some minor differences,
-
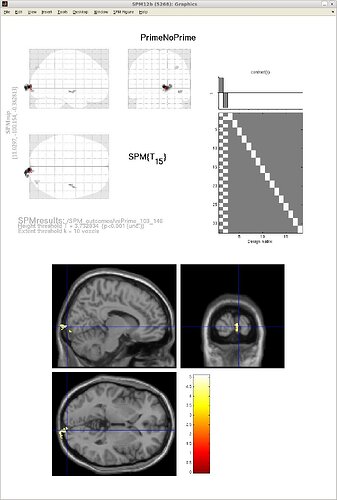
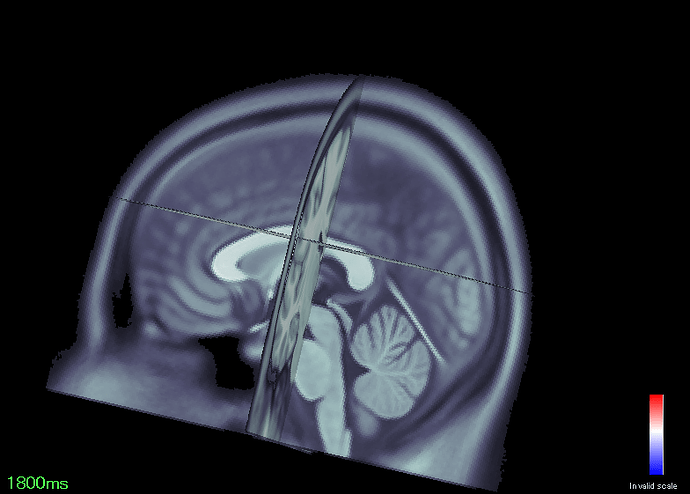
For this same test, an effect is located near the occipital cortex, but actually outside of it (better visualized in the second figure attached),
It seem that all my volumes are displaced posteriorly relative to the SPM template,
Any hint is wellcome,
José Luis
Hi Jose Luis,
It looks like the brain you are displaying here is from a SPM normalized MRI.
If you want the sources calculated in Brainstorm to match exactly the anatomy in the SPM view, you have to use the same MRI volume as the one used in Brainstorm, as illustrated here:
http://neuroimage.usc.edu/brainstorm/ExportSpm8#Display_the_results
To export the structural MRI volume from Brainstorm: right-click on the MRI > File > Export MRI > File format = NIfTI.
Cheers,
Francois
Hello Francois,
Ok, it looks better, but how should I do to get the coordinates relative to the brainstorm template?,
Thanks,
José Luis
I’m not sure but I think the coordinates are all in Brainstorm “MNI coordinates” (last line in the MRI Viewer).
SPM processes whatever you give it in input and do not normalize anything unless you ask for it explicitly.
It’s just the display of the background image of the SPM glass brain that is wrong.
(I’m not 100% sure of what I claim here, I encourage you to double-check it manually)
Hello Francois,
Sorry, I have to come back with same question again,
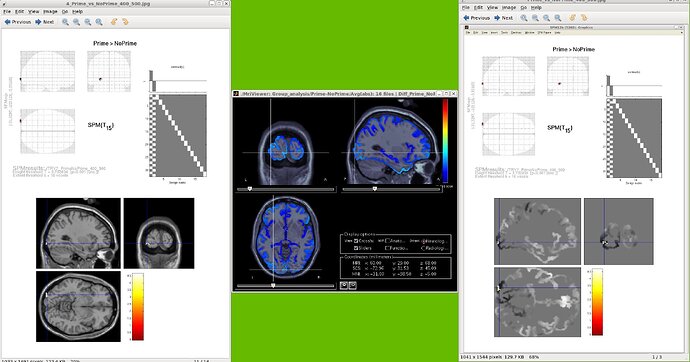
I can visualize my results correctly using a BS template,
However, I'm still having problems to get the coordinates. You can see in the figure that the x (-31) and z (-5) coordinates seem prett well. However, while SPM is telling me that the y coordinate is -103, for BS this coordinate seem to be rather -98, I would like to know if there a way to know with a reasonable precision the values of these coordinates,
thanks
José Luis
Hi Jose Luis,
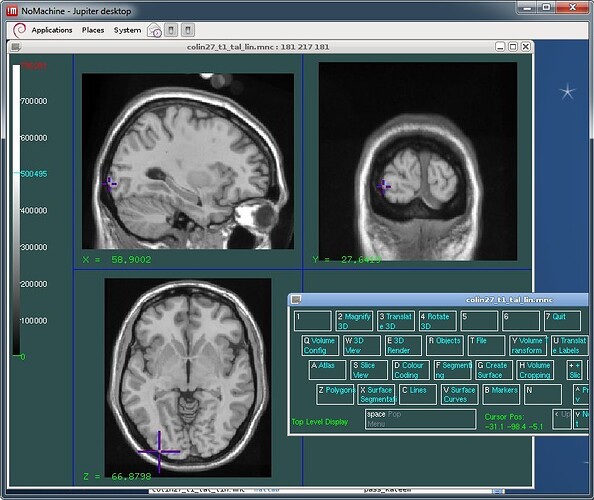
I double-checked the coordinates of the slices you show in the Brainstorm figure, and the coordinates are correct in Brainstorm, wrong in your SPM figure.
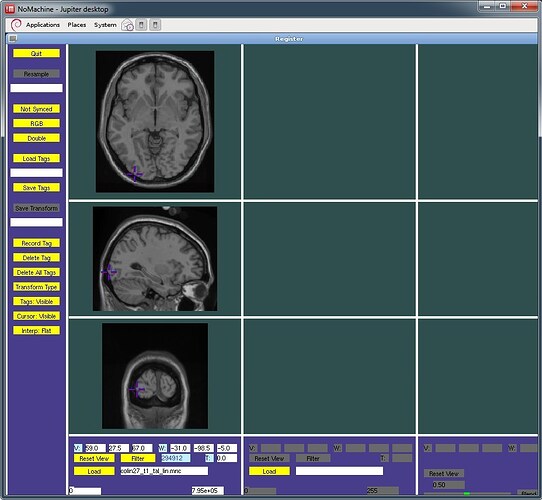
I checked with two of the MNI display tools (Register and Display), using the reference Colin27 volume (colin27_t1_tal_lin.mnc) downloaded from the MNI website:
http://www.bic.mni.mcgill.ca/ServicesAtlases/Colin27
See attached screen captures. For the same MNI coordinates ("world coordinates" in MNI environment), you will notice a difference of 1 voxel between what is shown in voxel coordinates between Brainstorm and the MNI tools because of a different way to index the volumes. Except for that, they validate the Brainstorm display.
I'm not sure why your are getting wrong coordinates in SPM.
Probably because you are using the NIFTI volumes generated with Brainstorm. In these volumes, I do not save at any referential, just the volume itself.
I guess SPM has no way to know what the referential is, and sets a random (and wrong) origin in the volume.
Can you work in voxel coordinates in SPM, instead of those wrong "world coordinates" ? This would be more reliable to exchange position information between Brainstorm and SPM.
Alternatively, you can ask the SPM developers to fix their code to display correct "MNI coordinates" in your case.
Francois
Ok,
I’ll try to see it at the SPM forum,
José Luis
Hello José, and or Francois,
Don’t know if you still frequent this forum as your post was from three years ago. But, did this issue ever get resolved? I also want to do analyses with Brainstorm generated images in SPM8.
Is there a thread regarding using exported nii files in SPM8 somewhere?
I am trying to run a paired t test on exported NIFTI volumes generated with Brainstorm.
A head model was made based on the MRI volume.
An MNI current density, unconstrained, source estimation kernel was calculated.
Source files for each trial were then averaged over time, and then average across trials to retain non-phase locked activation (i.e. what SPM refers to as an “induced” analysis).
This process was done for two different timepoints (i.e. stimulus response, and baseline).
This process produced two image files per subject (i.e. average activity during stimulus response across trials, and average activity during baseline across trials).
I exported these files using the file -> spm8 volume
I am using the template brain, ICBM152 for these analyses.
Digitized headpoints were corregistered with the template using the warp procedure in Brainstorm.
I also exported the T1 of the ICBM152 template in the default anatomy folder as a NIFTI file and put it in the same folder as the other NIFTI files I created.
I ran SPM8 and attempted to perform a paired t test.
However, when I try to generate the SPM analysis matrix, I get an error saying there is a “dimension mismatch”.
I suspect there must be some additional procedure I must do to ensure that the dimensions of all my image files are the same? Does anyone have any idea what that might be?
I observed that the file sizes of the exported NIFTI image files are different depending on the subject. I don’t understand why this is happening, especially when the information should all be averaged to a single timepoint.
Importantly, when I attempt to run a parametric paired t test in Brainstorm with the image files in A and B, the output also says that the matrices are different sizes.
I really appreciate any help anyone can offer. Thank you.
Jared
P.S. The stimulus response period and the baseline period are not the same length of time. Opening up the files in the Matlab workspace reveals that the matrix dimensions of both the stimulus response and baseline images within any given subject are the same, except for the time matrix. Also, the low and high values for the ImageGridAmp are not the same. I presume other matrices might also be different between subjects due to differences in things such as the number of bad trials.
Hello,
Have you followed the instructions in this tutorial?
http://neuroimage.usc.edu/brainstorm/ExportSpm8
The size of the volumes may differ between subjects if you selected the option “Cut empty slices” in the process options.
If you’re running the t-test within Brainstorm, you must explicitly indicate you want to average over time, otherwise it will run a t-test separately for each time sample, and if the number of time samples is not equal for all the samples, it will crash.
If you have error with Brainstorm, please post the full error messages.
Cheers,
Francois
PS: We have modified the .nii import/export functions in the past few weeks.
Make sure you update Brainstorm before trying to export again your volumes.
Hello Francois,
Thank you so much always for your replies. I am running the latest version of Brainstorm.
As I said, my source files were averaged over time for each trial, then averaged across trials.
I used two different time periods within each trial for my conditions (Baseline and Stimulus Response).
Because I averaged over time, I would expect the source files to have the same dimensions. But that does not appear to be the case.
When I run a parametric t test within Brainstorm (with the option "average selected time window" selected), I get the following warning:
Error: File #2 contains a data matrix that has a different size:
Error: File #3 contains a data matrix that has a different size:
Error: File #4 contains a data matrix that has a different size:
Error: File #5 contains a data matrix that has a different size:
Error: File #6 contains a data matrix that has a different size:
Error: File #7 contains a data matrix that has a different size:
Error: File #8 contains a data matrix that has a different size:
Error: File #9 contains a data matrix that has a different size:
Error: File #10 contains a data matrix that has a different size:
Error: File #11 contains a data matrix that has a different size:
Error: File #12 contains a data matrix that has a different size:
Error: File #13 contains a data matrix that has a different size:
Error: File #14 contains a data matrix that has a different size:
And the resulting t-test map file, though produced, has an invalid scale and shows no results.
As for SPM, your comment about the option, "Cut empty slices" was indeed a problem. I am currently re-exporting the files now and they all appear to be the same size. I will write again soon with the results of another test with SPM. However, it's a shame that the t-test doesn't seem to work in Brainstorm. Ideally, I wanted to compare the results from both software types.
Anyway, more soon. Thank you!
Jared
Hi Francois,
OK. Well, all my NIFTI files are the same size now. However, SPM still is not happy. I get this error message when running the paired t test in SPM:
'Failed 'Factorial design specification’
エラー ==> spm_check_orientations at 54
The orientations etc must be identical for this procedure.
In file “C:\Users\meglab\Documents\MATLAB\spm8\spm_check_orientations.m” (v4199), function “spm_check_orientations” at line 54.
In file “C:\Users\meglab\Documents\MATLAB\spm8\config\spm_run_factorial_design.m” (v4537), function “spm_run_factorial_design” at line 641.
The following modules did not run:
Failed: Factorial design specification’
I know you are not an expert at SPM, so I don’t expect you to know what this means. But perhaps you have some insight on how Brainstorm might be creating image files with different orientations? Could this be due to each subject being warped to the template brain? Do I need to extrapolate sources from warped images back onto the original template?
Thank you so much for your help,
Jared
In Brainstorm: If you are using a volume source model, computed independently for each subject, you may get different number of source points across subjects, and this would prevent you from averaging/comparing them together. This is described in these tutorials:
http://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource#Group_analysis
http://neuroimage.usc.edu/brainstorm/Tutorials/CoregisterSubjects#Volume_source_models
You shouldn’t have this problem with surface source models, as all the warped surfaces should always have the same number of vertices.
In SPM: I don’t know what the problem can be here.
If you have exported all the files using the same options (as illustrated here: http://neuroimage.usc.edu/brainstorm/ExportSpm8), all the files should have the same dimensions. I don’t know what SPM is not happy about here.
This would require some debugging. You can place a breakpoint in spm_check_orientations.m, line 54 and see what the problem is.
Any particular reason why you are using spm8 and not SPM12?
Can you try with SPM12 to see if you are getting the same issues?
Dear Francois,
Thank you so much always for your kind and informative replies.
It was the weekend and we had a scheduled power shutdown at our institution yesterday, so I’m just getting to this now.
I will read the Brainstorm tutorial info you linked. Thank you very much.
And, thank you for your advice regarding SPM8. I’ll see about putting in the breakpoint you mentioned. I’m using SPM8 because the data I’m processing was previously analyzed using it. We are reanalyzing it now after cleaning the data with Brainstorm. Ideally, I had wanted to export the cleaned data prior to source estimation for use in SPM8. I’ve noticed that some other people have also been interested in this. However, it appears to be not so easy to do.
I am now reprocessing with surface source models, and will try with SPM12. I will let you know the results here.
Thank you again,
Jared
Dear Francois,
It has been very busy. I wanted to let you know that your advice on this was great! We were able to successfully export and analyze our source level data on SPM12. I have yet to try and get the process to work for SPM8 analyses, but I have little motivation to do so now as I think surface models are best for our studies at present.
Anyway, thank you again!
Best,
Jared