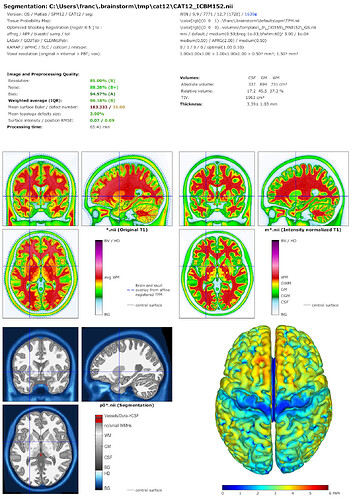
Hello François,
I post my question here because I have similar errors. It has been several weeks now that I am trying to segment my own MRIs with CAT12 on Brainstorm as you suggested :
- Create a new subject (Default anatomy: NO, Default channel file: NO)
- Right-click on the subject > Import MRI (from any file format, no need to run FreeSurfer before, any .nii or event DICOM would work)
- Right-click on the MRI > CAT12 MRI segmentation
But I always get the same issue, the error message you can see below. I tried to run CAT12 independently and I had another error when I tried to segment MRI. I think this may be a CAT12 issue and I was wondering if you have any idea how to resolve it?
Thank you in advance for your reply!
To help you identify the problem, I am posting everything that appeared in the MATLAB command window including the error below:
SPM12: spm_cat12 (vCAT12.9) 10:23:18 - 08/04/2024
========================================================================
_______ ___ _______
| ____/ / _ \ \_ _/
| |___ / /_\ \ | | Computational Anatomy Toolbox
|____/ /_/ \_\ |_| CAT12.9 - https://neuro-jena.github.io
CAT default file:
C:\Users\T4-Admin\.brainstorm\plugins\cat12\cat12\cat_defaults.m
Item 'Tensor Calculation', field 'prog': Value must be a function or function handle on MATLAB path.
Item 'Probabilistic Tracking', field 'prog': Value must be a function or function handle on MATLAB path.
Item 'Mori Streamline Tracking', field 'prog': Value must be a function or function handle on MATLAB path.
Item atlas: No field(s) named
warped
dartel
------------------------------------------------------------------------
08-Apr-2024 10:23:25 - Running job #1
------------------------------------------------------------------------
08-Apr-2024 10:23:25 - Running 'CAT12: Segmentation'
------------------------------------------------------------------------
CAT12.9 r2560: 1/1: .\.brainstorm\tmp\cat12_240408_102313\P04-PE.n
------------------------------------------------------------------------
SANLM denoising (medium) Your version of CAT12 is up-to-date.
29s
Affine preprocessing (APP)
Initialize 7s
Estimate background 3s
Initial correction 8s
Refine background 3s
Final correction 5s
Final scaling 4s
34s
Correct center-of-mass 3s
Affine registration 7s
SPM preprocessing 1 (estimate 1 - TPM registration): 54s
SPM preprocessing 1 (estimate 2): 110s
SPM preprocessing 2 (write)
Write Segmentation 22s
Update Segmentation 14s
Update Skull-Stripping 18s
Update probability maps 3s
57s
Global intensity correction 7s
SANLM denoising after intensity normalization (medium) 3s
Fast Optimized Shooting registration 2s
39s
Local adaptive segmentation (LASstr=0.50)
Prepare maps 3s
Prepare partitions 2s
Prepare segments (LASmod = 1.00) 6s
Estimate local tissue thresholds (WM) 16s
Estimate local tissue thresholds (GM) 21s
Intensity transformation 0s
SANLM denoising after LAS (medium) 21s
71s
ROI segmentation (partitioning)
Atlas -> subject space 8s
Major structures 9s
Ventricle detection 4s
Blood vessel detection 5s
WMH detection (WMHCstr=0.50 > WMHCstr'=0.00) 19s
Manual stroke lesion detection 0s
Closing of deep structures 1s
Side alignment 2s
Final corrections 1s
50s
Apply enhanced blood vessel correction 1s
Amap using initial SPM12 segmentations (MRF filter strength 0.05) 101s
AMAP peaks: [CSF,GM,WM] = [0.36±0.06,0.68±0.07,0.98±0.05]
Final cleanup (gcutstr=0.25)
Level 1 cleanup (ROI estimation) 2s
Level 1 cleanup (brain masking) 1s
Level 2 cleanup (CSF correction) 1s
Level 3 cleanup (CSF/WM PVE) 1s
5s
2s
Write result maps 21s
Surface and thickness estimation
lh:
Thickness estimation (0.50 mm³) 46s
Create initial surface 22sWarning: [GIFTI] Parsing of XML file C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\surf\lh.layer4x.P04-PE.gii failed.
> In read_gifti_file (line 17)
In gifti (line 108)
In cat_surf_fun>cat_surf_evalCS (line 1295)
In cat_surf_fun (line 200)
In cat_surf_createCS2 (line 1008)
In cat_main1639 (line 812)
In cat_run_job1639 (line 1296)
In cat_run_newcatch (line 40)
In cat_run>run_job (line 1271)
In cat_run (line 757)
In cfg_run_cm (line 29)
In cfg_util>local_runcj (line 1717)
In cfg_util (line 972)
In spm_jobman>fill_run_job (line 469)
In spm_jobman (line 247)
In process_segment_cat12>Compute (line 356)
In process_segment_cat12>ComputeInteractive (line 440)
In process_segment_cat12 (line 28)
In bst_call (line 28)
In tree_callbacks>@(h,ev)bst_call(@process_segment_cat12,'ComputeInteractive',iSubject,iAnatomy) (line 3110)
Topology correction: Warning: Escaped character '\U' is not valid. See 'doc sprintf' for supported special characters.
> In cat_io_cprintf (line 335)
In cat_run_newcatch (line 57)
In cat_run>run_job (line 1271)
In cat_run (line 757)
In cfg_run_cm (line 29)
In cfg_util>local_runcj (line 1717)
In cfg_util (line 972)
In spm_jobman>fill_run_job (line 469)
In spm_jobman (line 247)
In process_segment_cat12>Compute (line 356)
In process_segment_cat12>ComputeInteractive (line 440)
In process_segment_cat12 (line 28)
In bst_call (line 28)
In tree_callbacks>@(h,ev)bst_call(@process_segment_cat12,'ComputeInteractive',iSubject,iAnatomy) (line 3110)
------------------------------------------------------------------------
CAT Preprocessing error for P04-PE:
------------------------------------------------------------------------
Surface file C:Warning: Escaped character '\%' is not valid. See 'doc sprintf' for supported special characters.
> In cat_run_newcatch (line 175)
In cat_run>run_job (line 1271)
In cat_run (line 757)
In cfg_run_cm (line 29)
In cfg_util>local_runcj (line 1717)
In cfg_util (line 972)
In spm_jobman>fill_run_job (line 469)
In spm_jobman (line 247)
In process_segment_cat12>Compute (line 356)
In process_segment_cat12>ComputeInteractive (line 440)
In process_segment_cat12 (line 28)
In bst_call (line 28)
In tree_callbacks>@(h,ev)bst_call(@process_segment_cat12,'ComputeInteractive',iSubject,iAnatomy) (line 3110)
1809 - loadSurf
1072 - cat_surf_createCS2
812 - cat_main1639
1296 - cat_run_job1639
40 - cat_run_newcatch
1271 - run_job
757 - cat_run
29 - cfg_run_cm
1717 - local_runcj
972 - cfg_util
469 - fill_run_job
247 - spm_jobman
356 - Compute
440 - ComputeInteractive
28 - process_segment_cat12
28 - bst_call
3110 - @(h,ev)bst_call(@process_segment_cat12,'ComputeInteractive',iSubject,iAnatomy)
------------------------------------------------------------------------
Error:cat_io_report:print: Error printing CAT error report.
Warning: MATLAB was unable to resolve the URL and open it in the browser:
MATLAB:nonLogicalConditional
> In web>displayWarningMessage (line 172)
In web (line 114)
In cat_io_senderrormail (line 49)
In cat_run_newcatch (line 223)
In cat_run>run_job (line 1271)
In cat_run (line 757)
In cfg_run_cm (line 29)
In cfg_util>local_runcj (line 1717)
In cfg_util (line 972)
In spm_jobman>fill_run_job (line 469)
In spm_jobman (line 247)
In process_segment_cat12>Compute (line 356)
In process_segment_cat12>ComputeInteractive (line 440)
In process_segment_cat12 (line 28)
In bst_call (line 28)
In tree_callbacks>@(h,ev)bst_call(@process_segment_cat12,'ComputeInteractive',iSubject,iAnatomy) (line 3110)
Please send mail to vbmweb@gmail.com and copy content of C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\report\cat_P04-PE.xml and C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\report\catlog_P04-PE.txt into mail if not already done or attach these files.
Remove "C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\label\catROI_P04-PE.xml" from dependency list because it does not exist!
One or multiple files do not exist and were removed from the dependency list
and following batches will may not work correctly!
Remove "C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\report\catreport_P04-PE.pdf" from dependency list because it does not exist!
One or multiple files do not exist and were removed from the dependency list
and following batches will may not work correctly!
Remove "C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\report\catreportj_P04-PE.jpg" from dependency list because it does not exist!
One or multiple files do not exist and were removed from the dependency list
and following batches will may not work correctly!
Remove "C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\surf\lh.central.P04-PE.gii" from dependency list because it does not exist!
One or multiple files do not exist and were removed from the dependency list
and following batches will may not work correctly!
Remove "C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\surf\lh.sphere.P04-PE.gii" from dependency list because it does not exist!
One or multiple files do not exist and were removed from the dependency list
and following batches will may not work correctly!
Remove "C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\surf\lh.sphere.reg.P04-PE.gii" from dependency list because it does not exist!
One or multiple files do not exist and were removed from the dependency list
and following batches will may not work correctly!
Remove "C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\surf\lh.thickness.P04-PE" from dependency list because it does not exist!
One or multiple files do not exist and were removed from the dependency list
and following batches will may not work correctly!
Remove "C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\surf\rh.central.P04-PE.gii" from dependency list because it does not exist!
One or multiple files do not exist and were removed from the dependency list
and following batches will may not work correctly!
Remove "C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\surf\rh.sphere.P04-PE.gii" from dependency list because it does not exist!
One or multiple files do not exist and were removed from the dependency list
and following batches will may not work correctly!
Remove "C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\surf\rh.sphere.reg.P04-PE.gii" from dependency list because it does not exist!
One or multiple files do not exist and were removed from the dependency list
and following batches will may not work correctly!
Remove "C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\surf\rh.thickness.P04-PE" from dependency list because it does not exist!
One or multiple files do not exist and were removed from the dependency list
and following batches will may not work correctly!
Remove "C:\Users\T4-Admin\.brainstorm\tmp\cat12_240408_102313\surf\rh.pbt.P04-PE" from dependency list because it does not exist!
One or multiple files do not exist and were removed from the dependency list
and following batches will may not work correctly!
08-Apr-2024 10:37:30 - Done 'CAT12: Segmentation'
Item 'Central Surfaces', field 'val': Number of matching files (0) less than required (1).
08-Apr-2024 10:37:30 - Failed to update inputs for 'Extract additional surface parameters'
Error using harvest
Dependencies resolved, but not suitable for this item.
In item Central Surfaces:
Dependency 1: CAT12: Segmentation (current release): Left Central Surface (out(1).lhcentral(:))
In file "harvest.m" (v5678), function "harvest" at line 47.
No executable modules, but still unresolved dependencies or incomplete module inputs.
The following modules did not run:
Skipped: Extract additional surface parameters
***************************************************************************
** Error: Line 0: MATLABbatch system
** Job execution failed. The full log of this run can be found in MATLAB command window, starting with the lines (look for the line showing the exact #job as displayed in this error message)
** ------------------
** Running job #1
** ------------------
**
**
** Call stack:
** >.m>MATLABbatch system at 0
**
***************************************************************************