"For the volume regions, you need to create a “Volume atlas” and create manually a scout for each region you want to use (not applicable for what you described here, but this way you will understand why not all the different ROIs are handled in the same way)" this is from another post. I am having trouble creating the volume atlas because I can hardly select voxels and not being able to make it bigger. How could I do that? Sorry if this sounds too naive.
|error|[link|NewSubject/@default_study/results_sLORET...](file://link|NewSubject/@default_study/results_sLORETA_EEG_KERNEL_210228_1239.mat|NewSubject/@rawmindMonitor_2021-02-17--13-02-13_pre_1/data_0raw_mindMonitor_2021-02-17--13-02-13_pre_1.mat)|Scout "Accumbens L" is not included in the source model.
If you use this region as a volume, create a volume scout instead (menu Atlas > New atlas > Volume scouts).|
| --- | --- | --- | --- | --- |.
ALL I SEE IS THIS ERROR WHEN COMPUTING TEST STATISTIC
I am having trouble creating the volume atlas because I can hardly select voxels and not being able to make it bigger
Mixed head models are very difficult do deal with for advanced post-processing, like statistics or connectivity analysis. Additionally, comparing signals obtained from different regions is difficult, as different subcortical regions use different source constraints.
An easier approach, at least as a first attempt, would be to use a volume source model.
https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource#Volume_scouts
(this procedure is also valid for creating volume scouts in mixed head models)
It is difficult to understand what you are trying to do here, and what is working or not working. If you need further help, include full error messages, screen captures showing the files in the database and their contents, a description of what you are trying to do...
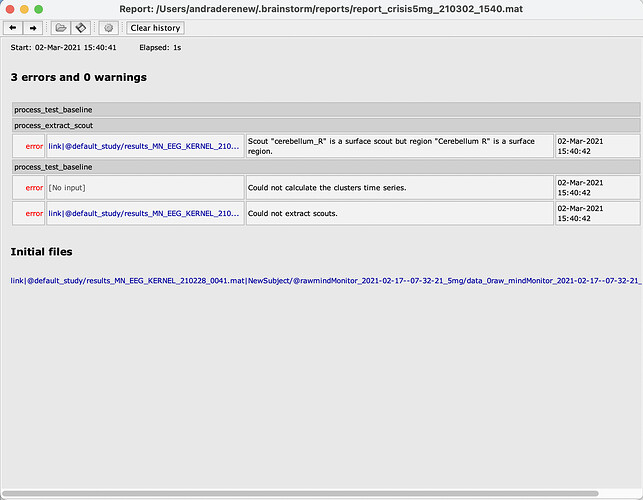
These are the screnshots after having created the volume scouts atlas. And trying to compute the statistics.
I fixed the error message you get here , which does not make sense:
Now you should get:
Scout "cerebellum_R" is a volume scout but region "Cerebellum R" is a surface region
In your forward model, you used the cerebellum as SURFACE. To extract some activity from it, you need to create a SURFACE scout.
If this does not seem clear to you, I recommend you work only with surface or volume head models. These mixed head models are very confusing, with high risks of manipulation errors...
So trying to carry on with my original idea. What I have to do is compute the volume head model and then create the volume scout atlas? is that correct?