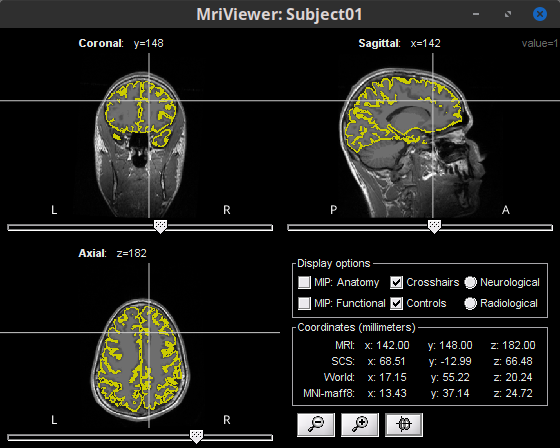
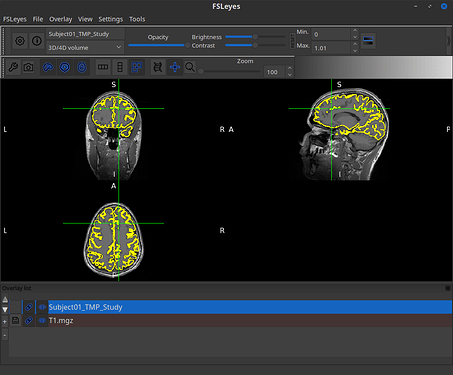
Hello experts, I want to know how to save the vertices used for head model to 3D volume format ?
The this 3D information is located in the GridLoc field in the HeadModel file:
https://neuroimage.usc.edu/brainstorm/Tutorials/HeadModel#On_the_hard_drive
What do you mean by saving them as a "3D volume format"?
Oh, very grateful for your reply! I mean is it possible to save this vertex information as a nii.gz file? I want to trace the fibers from these vertices.
That's a nice question. It is not possible to directly export the vertices as NIfTI file (nii.gz).
However, it is possible to interpolate those vertices to the (MRI) volume space, and then to save as NIfTI.
Try the script providing your surface to export as NIfTI and directory for the NIfTI file.
The script creates a temporary source file with all vertices set to 1; then, this surface sources are interpolated to the MRI volume and exported as NIfTI. Finally the temporary files are deleted.
The result is the same as the one that you can observe by right-clicking the cortex, then MRI registration > Check MRI/surface registration...
Oh, very grateful for your reply, I will try to run this script, thank you ![]() !
!
I tried it, it is just what I want, very thanks!
I have another question for this, how to know the mapping between the surface vertices and the NIFTI files?
In Brainstorm, the interpolation from a surface to the MRI volume is computed with the function tess_tri_interp:
Ok, I see, thank you!
Update. This can be done now by creating a surface scout that comprises the entire cortex (e.g., merging the hemisphere scouts in the Structures atlas), and then exporting the surface scout as MRI mask. See this other thread: