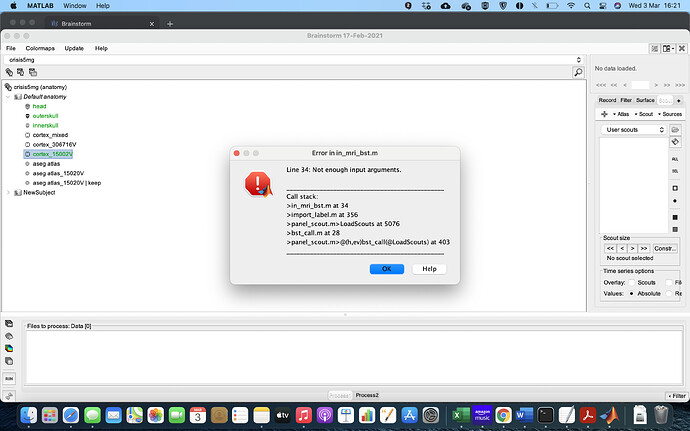
I cannot import the atlas. I do Atlas> Load Atlas and I get the error I show in screenshot after saying yes to the transformation of the volume.
I must admit that if I try doing the same but in another protocol there is no problem. Seems to me that mixing up many things (different headmodels, etc) in the same protocol leads to some sort of errors.
I opened a new protocol and did the mixed headmodel so I obtained the inversed current density unconstrained map. Then I make a new atlas volume scouts add the scouts to the atlas from aseg.mgz. So my issue now is that when I try to run either connectivity or Test I get this error. No matter how many scouts are there. I have tried with smaller and smaller numbers each time.
- Do not maximize the Brainstorm window, in order to see what's behind:
https://neuroimage.usc.edu/brainstorm/Tutorials/CreateProtocol?highlight=(maximize)#Main_interface_window - Start by following all the introduction tutorials (section Get started), using the example dataset, before trying to process your own data, this will save you a lot of trial-and-errors:
https://neuroimage.usc.edu/brainstorm/Tutorials - It looks like you have deleted the MRI in the default anatomy, you anatomy folder is all messed up: delete your protocol and start over. Never modify the default anatomy template, modify only the subject files
So my issue now is that when I try to run either connectivity or Test I get this error.
I need mode information in order to reproduce your error. Please post screen captures showing:
- Your database: anatomy files you are using
- 3D figure showing your 3D scouts
- 3D figures showing the anatomical elements you used for the computation of your head model
- Your database: functional files (channel file, head model, inverse model)
- 3D figures showing the source results
- Screen capture of the option of the process that doesn't work
these are almost all the screenshots but number 3 I hope this helps to clarify
What I have done in this case is add scout to atlas in source from freesurfer aseg+aparc.mgz having merged 15000 vertices from cortex and aseg. I added to all scouts being surface and constrained. Also for head modeling computation I did custom source. And current density for source estimation.
I'm trying to compute connectivity or test statistic as in the tutorials. In the screenshots I compute correlationNxN.
It looks like you've been trying to import an individual volume parcellation (eg. the aseg+aparc.mgz) as an surface atlas on the template brain. But these tools are not designed to work this way. In your screen capture with the scouts, it's quite obvious that this did not work at all.
I don't think these mixed head models are a good solution for you.
Since you don't seem to be clear on the types of files you are manipulating, I recommend you start with something simpler: either a surface head model (in that case, you already have the FreeSurfer parcellations available in the Scout tab), or a volume head model (in that case, you may import a MGZ file as a volume atlas, but only for the same subject).
Keep your analysis simple. Making your source analysis more complicated will only increase your chances of making mistakes.
If you have MGZ files, I guess you have individual data processed with FreeSurfer. You should import these before processing your data.
And once more, I insist on the importance of starting by the following the introduction tutorials before trying to process personal data.
Yes indeed I have been importing aseg+aparc.mgz I didn't know they were volume parcellation files. Now that I know I will try to run as volume atlas. That I guess will allow me to work with cerebellum. Which is essential to what I am doing. Please let me know if this is feasible. I cannot work just with cortex. If it is harder to work with mixed models. I will have to learn that. It is not an option to work just with cortex in my case. About FreeSurfer I am using fsaverage. Although I do know how to work with FreeSurfer, FSL, and SPM as well.
If you feel it is possible to work on a tutorial to achieve what I need(all regions of the brain to compute source reconstruction, connectivity, test, difference in 2 runs). Please let me know how can I help. You would just need to guide me and review the tutorial in case of errors. I am willing to learn and help this project for all those in the same situation I am be able to work with this.
That I guess will allow me to work with cerebellum. Which is essential to what I am doing. Please let me know if this is feasible.
https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource
About FreeSurfer I am using fsaverage
I don't recommend using the fsaverage subject in Brainstorm, use the ICBM152 template instead.
Do not mix ICBM152 and fsaverage files, these are two atlases that are not directly compatible.
In the tutorial you mention is not clear to me what are the steps for the whole process. May be it is in different tutorials so let me clarify what I do to check if there is some mistake.
I combine aseg with cortex and obtain a whole brain(without dba or source of any type, it is not clear here if I have to add something with volume how??). Then I compute headmodel, covariance matrix and source for mri volume option. Here I see I think is where we add volume scouts from atlas. I can do that but I already have a source model where almost there is activation for each time in the whole brain. May be I did something wrong.
About fsaverage I didn't meant to use it as template but I use parcellations from those files in fsaverage. If not where from I can obtain aseg+aparc files? From a segmentation with recon-all that is computing freesurfer over IBCM152 template. Where can I find this template then? To my knowledge I can download from a link of McGill but it doesn't seem to work. May be it is dowloaded in another software as well where I can get it easily.
Also I want to mention that when I use ay atlas from spm to scouts I ony see numbers and not the names of those scouts. Is there something I am missing?
I combine aseg with cortex and obtain a whole brain
Extract only the cerebellum, if you are interested only with the cerebellum.
Since it looks like you are working with an anatomy template: download the ICBM152_2019 full template (right-click on the anatomy folder > Use template > MNI > ...), it includes a surface with cortex+cerebellum.
Then I compute headmodel, covariance matrix and source for mri volume option. Here I see I think is where we add volume scouts from atlas. I can do that but I already have a source model where almost there is activation for each time in the whole brain.
Create a volume atlas, create a few scouts, make sure you can do what you want with these scouts.
As a second step, you may try to import a MNI template as an atlas (in MNI space, therefore not from fsaverage).
We've added the automatic download for many MNI templates in Brainstorm, but we can't use them yet as volume atlases. Coming soon...
About fsaverage I didn't meant to use it as template but I use parcellations from those files in fsaverage. If not where from I can obtain aseg+aparc files? From a segmentation with recon-all that is computing freesurfer over IBCM152 template.
Exactly.
This will be available in a few weeks (or months) easily in Brainstorm. But for now, you need to process the volume yourself.
If you want to use the FreeSurfer Destrieux or DK/DKT40 atlases, I'd recommend you use the surface version (and therefore a surface head volume) - then they are already available in the cortex surfaces of the template.
Also I want to mention that when I use ay atlas from spm to scouts I ony see numbers and not the names of those scouts. Is there something I am missing?
The volume parcellations you import contain only integers. These are the values you see.
We are in the process of adding default labels for some atlases, but this is not finished yet.