|
Size: 9054
Comment:
|
Size: 24652
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| <<TableOfContents(2,2)>> | ''Author: Francois Tadel'' This page describes the various coordinate systems used in Brainstorm, and explains how to convert beetween referentials. <<TableOfContents(3,2)>> |
| Line 5: | Line 9: |
| Brainstorm uses the CTF coordinate sytem. All the surfaces, sensors and additional points are converted into this system when they are imported in Brainstorm. | Brainstorm uses the CTF head coordinate system. All the surfaces, sensors and additional points are converted into this system when they are imported in Brainstorm. |
| Line 9: | Line 13: |
| * __Based on__: Nasion, left pre-auricular point (LPA), and right pre-auricular point (RPA). * __Origin__: Midway on the line joining LPA and RPA * __Axis X__: From the origin towards the Nasion (exactly through) * __Axis Y__: From the origin towards LPA in the plane defined by (NAS,LPA,RPA), and orthogonal to X axis * __Axiz Z__: From the origin towards the top of the head |
<<HTML(<div style="float: right; clear: left; margin: -20px 20px 0px 0px;">)>> {{attachment:scs_axes.png||width="220"}} <<HTML(</div>)>> * '''Based on''': Nasion, left pre-auricular point (LPA), and right pre-auricular point (RPA). * '''Origin''': Midway on the line joining LPA and RPA. <<BR>> * '''Axis X''': From the origin towards the Nasion (exactly through). * '''Axis Y''': From the origin towards LPA in the plane defined by (NAS,LPA,RPA), and orthogonal. to X axis. * '''Axiz Z''': From the origin towards the top of the head. <<BR>><<BR>><<BR>> The affine transformation from MRI to SCS coordinates is saved in the [[https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#On_the_hard_drive:_MRI|MRI SCS structure]]: * '''SCS.R''': [3x3] rotation matrix from MRI coordinates to SCS coordinates. * '''SCS.T''': [3x1] translation matrix from MRI coordinates to SCS coordinates. |
| Line 20: | Line 35: |
| {{attachment:NAS.gif}} | {{attachment:NAS.gif||width="599",height="213"}} |
| Line 23: | Line 38: |
| The proper definition of the preauricular point is: "a point of the posterior root of the zygomatic arch lying immediately in front of the upper end of the tragus". The problem with this definition is that it can be really difficult to localize precisely on the MRI. It can lead to severe misregistration between MRI and MEG/EEG. Our proposition is to use instead the junction between the tragus and the helix, marked with a red dot in the following figure. It can be located much more precisely both anatomically and on the MRI slices. {{attachment:preauricular.gif}} |
<<HTML(<div style="float: left; clear: right; margin: 0px 20px 0px 20px;">)>> {{attachment:preauricular.gif||width="148",height="235"}} <<HTML(</div>)>> The proper definition of the preauricular point is: "a point of the posterior root of the zygomatic arch lying immediately in front of the upper end of the tragus". This is illustrated with the green point below. The problem with this definition is that it can be really difficult to localize precisely on the MRI. It can lead to severe misregistration between MRI and MEG/EEG. More information on the anatomical location of this point on the [[http://fieldtrip.fcdonders.nl/faq/how_are_the_lpa_and_rpa_points_defined|FieldTrip website]]. Our proposition is to use instead the junction between the tragus and the helix, marked with a '''red dot''' in this figure. It can be located much more precisely both anatomically and on the MRI slices. You can choose to use this definition of the "preauricular point" or not. '''The most important is to use the same convention for all the steps: digitization of head points''' (eg. Polhemus) '''and analysis with Brainstorm'''. <<HTML(<div style="clear: both;"></div>)>> |
| Line 31: | Line 52: |
| {{attachment:LPA.gif}} You can choose to use this definition of the "preauricular point" or not. The most important is to use the same convention for all the steps: digitization of head points with a magnetic system (Isotrak Polhemus, ...), and analysis with Brainstorm.<<BR>><<BR>> |
{{attachment:LPA.gif||width="599",height="213"}} |
| Line 36: | Line 55: |
| Using a CTF system, you would probably prefer to use the head localization coils to indicate those NAS/LPA/RPA points, instead of those "anatomically correct" points. The coordinate system in the CTF files is based on the position of the three coils you stick on the head of the subject. Typically, the nose coil is slightly above the nasion, and the ear coils about one centimeter more frontal than the points that were previously described. | Using a CTF system, you would probably prefer to use the head localization coils to indicate these NAS/LPA/RPA points, instead of these "anatomically correct" points. The coordinate system in the CTF files is based on the position of the three coils you stick on the head of the subject. Typically, the nose coil is slightly above the nasion, and the ear coils about one centimeter more frontal than the points that were previously described. |
| Line 42: | Line 61: |
| __'''IMPORTANT'''__: Those comments __do not apply__ if you use the [[Tutorials/TutDigitize|Brainstorm Digitize]] program to drive the Polhemus device. In this case, you are asked to digitize two sets of NAS/LPA/RPA points: the coils and the real anatomical points. Just make a copy of the .pos file saved by Digitize in the .ds folder, then set the real anatomical points in the MRI. Brainstorm is going to handle automatically the transformation between the CTF coils coordinate system and the real subject coordinate system. The result is usually much more accurate and does not require to take pictures of the subjects. == Talaraich coordinates == The atlas that Jean Talairach established for the human brain is base on two points, the posterior and anterior commissure, and the midsagittal plane. The third point needed here, the interhemispheric point, is used to define this plane. To learn more about the Talairach coordinate system, please visit [[http://en.wikipedia.org/wiki/Jean_Talairach|this Wikipedia page]]. |
__'''IMPORTANT'''__: These comments __do not apply__ if you use the [[Tutorials/TutDigitize|Brainstorm Digitize]] program to drive the Polhemus device. In this case, you are asked to digitize two sets of NAS/LPA/RPA points: the coils and the real anatomical points. Just make a copy of the [[http://neuroimage.usc.edu/brainstorm/CoordinateSystems#Brainstorm_.pos_files|.pos file]] saved by Digitize in the .ds folder, then set the real anatomical points in the MRI. Brainstorm is going to automatically handle the transformation between the CTF coils coordinate system and the real subject coordinate system. The result is usually much more accurate and does not require to take pictures of the subjects. === Using a default anatomy === The fiducial points (Nasion, LPA, RPA) used in your recordings might not be the same as the ones used in the anatomy templates in Brainstorm (ICBM152, Colin27, FSAverage). By default, the LPA/RPA points are defined at the junction between the tragus and the helix, as represented with the red dot. If you want to use an anatomy template but you are using a different convention when digitizing the position of these points, you have to modify the default positions of the template with the MRI Viewer. * Go to the anatomy view * In (default anatomy), right-click on the MRI > Edit MRI * Modify the position of the fiducial points to match your own convention * Click on [Save], it will update the surfaces to match the new coordinate system == MRI coordinates == Coordinates system used in the space of the MRI volume, in millimeters: * __Axis X__: left to right * __Axis Y__: posterior to anterior * __Axis Z__: inferior to superior * The first voxel at the left-posterior-inferior corner of the MRI volume is indexed (1,1,1). The corresponding MRI coordinates of the center of this voxel in millimeters is (voxelsize_x, voxelsize_y, voxelsize_z). == World coordinates == Coordinates system used in the original MRI file, that was loaded as the reference MRI for a subject. It is defined by a 4x4 matrix saved directly in the MRI file. From NIfTI .nii files, it uses the matrix''' sform''' if available, or otherwise the matrix '''qform''' (see the [[https://nifti.nimh.nih.gov/nifti-1/documentation/nifti1fields/nifti1fields_pages/qsform.html|NIfTI documentation]]). From FreeSurfer .mgh/.mgz files, it uses the '''vox2ras''' matrix (see the [[https://surfer.nmr.mgh.harvard.edu/fswiki/CoordinateSystems|FreeSurfer documentation]]). In practical terms, it corresponds to the coordinates you see in the title bar when loading your volume in [[http://people.cas.sc.edu/rorden/mricron/index.html|MRIcron]], while the coordinates you see in the text boxes correspond in Brainstorm vocabulary to "voxel coordinates". For subject data, it corresponds most of the time to '''scanner coordinates'''. For atlases, it may correspond to normalized coordinates (e.g. MNI coordinates). If available, the transformation from voxels to world coordinates is saved in the field '''InitTransf{'vox2ras'}''' of the [[https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#On_the_hard_drive:_MRI|MRI structure]]. == MNI coordinates == In order to compare the anatomy of multiple human brains or compute group statistics, it is necessary to define a common reference system. Two types of normalized systems have been widely used in the literature in the past: the atlas defined by [[http://en.wikipedia.org/wiki/Jean_Talairach|Jean Talairach]], and the [[http://www.bic.mni.mcgill.ca/~louis/stx_history.html|MNI stereotaxic coordinates]]. You can read about the differences between these two systems [[http://imaging.mrc-cbu.cam.ac.uk/imaging/MniTalairach|here]]. While the Talaraich system has been progressively abandonned, the "MNI space" has been increasingly used, thanks to the development of automatic normalization methods in open-source neuroimaging software. Successive updates to the reference brain and software versions led to the coexistence of multiple "MNI spaces". For an exhaustive list, refer to these two websites: [[https://bids-specification.readthedocs.io/en/latest/99-appendices/08-coordinate-systems.html#standard-template-identifiers|BIDS specification]], [[https://www.lead-dbs.org/about-the-mni-spaces/|Lead-DBS]]. In Brainstorm, '''MNI coordinates''' refer to the normalized space defined in '''SPM12''', also referred to as '''IXI549''' '''space'''. Like the MRI coordinates above, it is in mm, and the (X, Y, Z) axes are oriented towards (right, anterior, dorsal). The origin is at the AC point (see Talairach section below) of the template, with the (negative) Y axis also going through the PC point of the template. Computing this MNI normalization for a subject does not alter the input MRI or create a new normalized volume: it only stores a transformation that allows referencing the subject's MRI volume with standardized MNI coordinates. There are two options: a linear and a non-linear approach. {{attachment:mni_norm.gif}} === Linear normalization === * A [4x4] affine transformation is computed and saved in the [[https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#On_the_hard_drive:_MRI|MRI NCS structure]]: * '''NCS.R''': [3x3] rotation matrix from MRI coordinates to MNI coordinates. * NCS.'''T''': [3x1] translation matrix from MRI coordinates to MNI coordinates. * The Brainstorm function [[https://github.com/brainstorm-tools/brainstorm3/blob/master/toolbox/anatomy/bst_normalize_mni.m|mri_normalize_maff.m]] is based on SPM12 function [[https://github.com/spm/spm12/blob/master/spm_maff8.m|spm_maff8.m]], described in [[http://www.ncbi.nlm.nih.gov/pubmed/15955494|(Ashburner 2005)]]. It is integrated in Brainstorm and does not require the installation of SPM12. * This transformation is fast to compute and to apply to 3D coordinates, and very light in terms of storage. It is appropriate for computing a deformation of the bounding box of the brain and adapted for quickly aligning brains in the same space, or for defining the seed of a [[https://neuroimage.usc.edu/brainstorm/Tutorials/TutVolSource#Volume_scouts|volume scout]]. However, it is not adapted to model the differences in the cortical folding of the individual brains or to the projection of a MNI parcellation to subject space. === Non-linear normalization === * Each voxel of the individual brain is projected to the MNI space independently from the others. In order to represent these non-linear deformation fields between volumes, we use the SPM12 representation: three volumes are saved per transformation, one for each dimension (X,Y,Z). One triplet of 3D matrices is necessary to convert from MNI to MRI coordinates (forward transformation), and one other to convert from MRI to MNI (inverse transformation). The following fields are stored in the [[https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#On_the_hard_drive:_MRI|MRI NCS structure]]: * NCS.'''iy''': 4D floating point matrix [x,y,z,3]: Inverse MNI deformation field. Same size as the Cube matrix, it gives for each voxel its coordinates in the MNI space, and is therefore used to convert from MRI coordinates to MNI coordinates. * NCS.'''y''': 4D floating point matrix [x,y,z,3]: Forward MNI deformation field. For some MNI coordinates, it gives their coorrespondance in the original subject space. To be interpreted, it has to be used with the matrix y_vox2ras. * NCS.'''y_vox2ras''': [4x4 double], transformation matrix that converts from voxel coordinates of the y volume to MNI coordinates. * NCS.'''y_method''': Algorithm used for the normalization (e.g. 'segment'=SPM12 Segment) * Example: The coordinates of the MRI voxel (i,j,k) correspond in MNI space to:<<BR>><<BR>> {{{ mni_x = NCS.iy(i,j,k,1); mni_y = NCS.iy(i,j,k,2); mni_z = NCS.iy(i,j,k,3); }}} * A non-linear transformation can represent the individual anatomical variations, and is adapted for warping an [[Tutorials/DefaultAnatomy#MNI_parcellations|MNI parcellation]] to the subject space. However, it is much heavier computationally. For each MRI volume, we need to store 6 additional volumes in the file, causing an important increase of the file size and the load time. The computation of MNI coordinates is also much longer, slowing down the interface of the MRI Viewer. * Non-linear MNI normalization is available in Brainstorm using the [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Anatomical_labelling|SPM12 Segment]] algorithm or the [[https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12|CAT12]] segmentation pipeline. Both SPM12 and CAT12 can be installed easily as [[https://neuroimage.usc.edu/brainstorm/Tutorials/Plugins|Brainstorm plugins]]. === References === * '''MNI space''': Refers to a system of coordinates for indexing the voxels within a volume, applicable to either individual MRI/CT volumes or anatomical atlases. * '''Anatomy template''': Set of anatomical files (MRI, surfaces, parcellations) representing a brain atlas. As the reference anatomy, we typically use the [[http://nist.mni.mcgill.ca/?p=904|ICBM125 2009c Nonlinear Asymmetric]], but other options are described in the tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/DefaultAnatomy|Using the anatomy templates]]. An anatomy template can be used for various purposes: * Replacing the subject anatomy in the Brainstorm database when no individual scans are available for the subject. Setting the anatomy of the subject is necessary for 2D/3D topography display and source estimation. * Warping an anatomy template to match the digitized head shape of a subject. See tutorial: [[Tutorials/TutWarping|Warping the anatomy templates]] * For group analysis at the source level: an intermediate step of projection on an anatomy template is necessary. See tutorial: [[Tutorials/CoregisterSubjects|Group analysis: Subjects coregistration]] * '''MNI template''': Anatomy template that has the particularity to be registered into an MNI space (MNI152, Colin27). * '''MNI parcellations''': Volume of integers where each value represents an anatomical label, and registered to an MNI space. A non-linear deformation field can be applied to an MNI parcellation in order to create a volume parcellation in subject space. The anatomical regions can be used as [[https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#Subcortical_regions:_Volume|volume scouts]], for reference in the [[https://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#Subcortical_regions:_Volume|MRI viewer]], or for [[https://neuroimage.usc.edu/brainstorm/Tutorials/ECoG|labelling SEEG contacts]]. See also tutorial [[https://neuroimage.usc.edu/brainstorm/Tutorials/DefaultAnatomy#MNI_parcellations|Using the anatomy templates]]. === MacOS troubleshooting === Error "'''mexmaci64 cannot be opened because the developer cannot be verified'''": * [[https://neuroimage.usc.edu/forums/t/error-on-macos-mexmaci64-file-cannot-be-opened-because-the-developer-cannot-be-verified/30159/2|Brainstorm forum]] * [[https://en.wikibooks.org/wiki/SPM/Installation_on_64bit_Mac_OS_(Intel)#macOS_Big_Sur|SPM12 website]] * [[https://www.fieldtriptoolbox.org/faq/mexmaci64_cannot_be_opened_because_the_developer_cannot_be_verified/|FieldTrip website]] == ACPC coordinates == This coordinate system is recommended for iEEG coordinates in the [[https://bids-specification.readthedocs.io/en/stable/appendices/coordinate-systems.html#ieeg-specific-coordinate-systems|BIDS specification]]. It relies on two points, the anterior and posterior commissures (AC and PC), and the midsagittal plane. The third point needed here, the interhemispheric point (IH), is used to define this plane. The anatomical landmarks are determined in the individual's anatomical scan and no scaling or deformations have been applied to the individual's anatomical scan. * __Origin__: Anterior commissure (AC) * __Axis X__: From left to right * __Axis Y__: Passing through PC and AC, from posterior to anterior * __Axis Z__: Along the mid-saggital plane (defined from AC, PC and IH), from inferior to superior References: * [[https://bids-specification.readthedocs.io/en/stable/appendices/coordinate-systems.html#ieeg-specific-coordinate-systems|BIDS specification]] * [[https://www.fieldtriptoolbox.org/faq/acpc/|FieldTrip page on ACPC]] and [[https://www.fieldtriptoolbox.org/faq/coordsys/#details-on-the-acpc-coordinate-system|FieldTrip page on coordinate systems]] * [[https://github.com/fieldtrip/fieldtrip/blob/master/utilities/ft_headcoordinates.m#L201|FieldTrip code]] * [[https://github.com/brainstorm-tools/brainstorm3/blob/master/toolbox/anatomy/cs_compute.m#L79|Brainstorm code]] |
| Line 58: | Line 161: |
| Talairach coordinates: (0, 0, 0). {{attachment:AC.gif}} |
{{attachment:AC.gif||width="601",height="214"}} |
| Line 71: | Line 172: |
| {{attachment:PC.gif}} | {{attachment:PC.gif||width="600",height="213"}} |
| Line 76: | Line 177: |
| {{attachment:IH.gif}} <<BR>><<BR>> == MNI coordinates == MNI coordinates are based on the same definition as the Talairach coordinates (AC/PC/IH) but using a different reference brain. You can read more about the differences between the references used in both cases on that page: http://imaging.mrc-cbu.cam.ac.uk/imaging/MniTalairach The default anatomy in Brainstorm (MNI/Colin27) is registered in the MNI space, therefore the AC/PC coordinates we get are directly in "MNI coordinates". When working on an individual anatomy, the coordinates indicated as "MNI" in the Brainstorm interface are the AC/PC coordinates, scaled linearly along the three axes to match the size of the Colin27 brain. __Warning__: We do not recommend using the "MNI coordinates" you see in Brainstorm on individual brains. If you want to get normalized coordinates, first project your source activations on the default anatomy:<<BR>> http://neuroimage.usc.edu/brainstorm/Tutorials/CoregisterSubjects |
{{attachment:IH.gif||width="599",height="213"}} <<BR>><<BR>> == Talairach coordinates == Similar to the ACPC coordinates, but with an additional piecewise scaling to match the Talairach-Tournoux (1988) atlas. See [[https://www.fieldtriptoolbox.org/faq/acpc/|FieldTrip documentation]]. No used in Brainstorm. == CapTrak coordinates == This coordinate system is accepted for EEG coordinates in the [[https://bids-specification.readthedocs.io/en/stable/appendices/coordinate-systems.html#eeg-specific-coordinate-systems|BIDS specification]]. It relies on the three head points LPA/RPA/NAS described above. * __Origin__: Between LPA and RPA * __Axis X__: From LPA through RPA exactly * __Axis Y__: Orthogonal to the X-axis through the nasion (NAS) * __Axis Z__: Orthogonal to the XY-plane through the vertex of the head References: * [[https://bids-specification.readthedocs.io/en/stable/appendices/coordinate-systems.html#eeg-specific-coordinate-systems|BIDS specification]] * [[https://www.fieldtriptoolbox.org/faq/coordsys/#details-of-the-captrak-coordinate-system|FieldTrip page on coordinate systems]] * [[https://github.com/brainstorm-tools/brainstorm3/blob/master/toolbox/anatomy/cs_compute.m#L111|Brainstorm code]] |
| Line 93: | Line 202: |
| * __Origin__: Intersection of the line L throught LPA and RPA and a plane orthogonal to L and passing throught the nasion. * __Axis X__: From the origin towards the RPA point (exactly throught) * __Axis Y__: From the origin towards the nasion (exactly throught) |
* __Origin__: Intersection of the line L through LPA and RPA and a plane orthogonal to L and passing through the nasion. * __Axis X__: From the origin towards the RPA point (exactly through) * __Axis Y__: From the origin towards the nasion (exactly through) |
| Line 99: | Line 208: |
| ==== MRI (voxels) to MRI (milimeters) ==== Get the structure sMri from the database: right-click on the MRI file > File > Export to Matlab > sMri. For one point P_mri(x,y,z), coordinates in voxels: . {{{ P_mm = P_voxels .* sMri.Voxsize; }}} For a set of points P_mri, size: [nPoints x 3]: . {{{ P_mm = bst_bsxfun(@times, P_voxels, sMri.Voxsize); }}} ==== MRI (meters) to SCS (meters) ==== You need the a structure sMri that contains at least the information about the fiducial points (nasion, left ear, right ear). Coordinates of the reference points in '''__millimeters__'''. . {{{ sMri.SCS.NAS = [x, y, z]; sMri.SCS.LPA = [x, y, z]; sMri.SCS.RPA = [x, y, z]; }}} To compute the transformation (rotation + translation) from the MRI coordinate system (milimeters) to the Brainstorm Subject CS (SCS, meters): . {{{ transf = cs_mri2scs(sMri); }}} In case the transformation fields are not available yet in the sMri.SCS structure: . {{{ sMri.SCS.R = transf.R; sMri.SCS.T = transf.T; sMri.SCS.Origin = transf.Origin; }}} To apply the transformation to a set of points P_mri(x,y,z), dimension P=[N,3], coordinates in __meters__: . {{{ P_scs = cs_mri2scs(sMri, P_mri' .* 1000)' ./ 1000; }}} Or (P_mri and P_scs still in '''__meters__'''): . {{{ P_scs = bst_bsxfun(@plus, (sMri.SCS.R * P_mri')', sMri.SCS.T' ./ 1000); }}} ==== MRI (voxels) to SCS (meters) ==== The successive transformations are: MRI (voxels) => MRI (millimeters) => SCS (millimteres) => SCS (meters) . {{{ P_mri = bst_bsxfun(@times, P_voxels, sMri.Voxsize); P_scs = cs_mri2scs(sMri, P_mri')' ./ 1000; }}} ==== SCS (meters) to MRI (voxels) ==== SCS (meters) => SCS (millimteres) => MRI (millimeters) => MRI (voxels) . {{{ P_mri = cs_scs2mri(sMri, P_scs' .* 1000)'; P_voxels = bst_bsxfun(@rdivide, P_mri, sMri.Voxsize); }}} == Feedback == <<EmbedContent(http://neuroimage.usc.edu/brainstorm3_register/get_feedback.php?Tutorials/CoordinateSystems)>> |
The conversions between coordinate systems is handled with one single function [[https://github.com/brainstorm-tools/brainstorm3/blob/master/toolbox/anatomy/cs_convert.m|cs_convert]]: * sMri: MRI structure from the database: right-click on the MRI file > File > Export to Matlab > sMri * src/dest: Source and destination coordinates systems: {'voxel', 'mri', 'scs', 'mni', 'world'} * P: List of points [Npoints x 3] * All the the coordinates have to be in '''meters''' (not millimeters). {{{ Pdest = cs_convert(sMri, 'src', 'dest', Psrc); }}} Examples: {{{ P_mri = cs_convert(sMri, 'voxel', 'mri', P_voxel); % Voxel => MRI coordinates P_mni = cs_convert(sMri, 'scs', 'mni', P_scs); % SCS => MNI coordinates }}} == Brainstorm .pos files == The .pos files generated with the [[Tutorials/TutDigitize|Brainstorm Digitizer]] contain both the positions of the '''anatomical landmarks''' (nasion, left ear, right ear) and the position of the '''head localization coils''' (HPI). Having the two sets of points allows us to convert automatically from the native CTF coordinates (based on the coils) to the recommended landmarks in Brainstorm (based on the anatomical landmarks). When a '''Brainstorm .pos file''' is present in the CTF '''.ds folder''', it is loaded automatically and used to convert the positions of the EEG and MEG sensors to the anatomical reference. This is done automatically without any message or user confirmation. In this case, you should mark the '''anatomical landmarks''' (real nasion and tragus/helix junctions) in the MRI Viewer when importing the anatomy. This is what is described in the [[Tutorials/ImportAnatomy|introduction tutorials]]. Otherwise, if you are importing CTF recordings '''without a Brainstorm .pos file''' in the .ds folders, or if you are importing the .pos files after linking the recordings to the database, you should typically select the positions of the '''CTF HPI coils''' in the MRI viewer. The syntax of these .pos files is the following, one line per digitized point: {{{ Number of EEG electrodes Index Label X Y Z : Defines an EEG electrode Index X Y Z : Defines a head shape point Label X Y Z : Defines a reference point }}} In the .pos files, the anatomical landmarks are labelled '''Nasion/LPA/RPA''', and the CTF HPI coils are labelled '''HPI-N/HPI-L/HPI-R''' (for nasion coil, left coil, right coil). Note that in the CTF software, the HPI coils are labelled NAS/LPA/RPA, which can lead to some confusion. We usually take multiple measures of each reference point and average them when we load the file. This helps improving the accuracy of the registration. {{{ 2 1 Cz 5.29886357 -0.39211620 13.97175211 2 Pz -2.72999896 0.56457819 14.80592908 3 9.47286768 0.09984190 -2.22056385 4 9.61745050 -0.00705346 -2.64471836 [...] 242 2.68917264 3.02778502 13.88281027 243 1.10525214 3.54043941 14.17347208 Nasion 9.59228188 0.08328658 -1.88504105 LPA -0.47469294 6.81987024 0.62870235 RPA -0.96029260 -6.72333418 0.16475241 Nasion 9.76823213 -0.11917776 -1.87417223 LPA -0.29274026 6.88415084 0.70120923 RPA -0.99183277 -6.70128616 0.08443862 HPI-N 10.61990713 0.01532629 0.00407281 HPI-L 0.20165313 6.74350937 -0.00134360 HPI-R -0.23124566 -6.76723759 -0.00008921 HPI-N 10.61095674 -0.01532629 -0.00407281 HPI-L 0.27017080 6.81335558 0.00134360 HPI-R -0.24057827 -6.78962736 0.00008921 }}} == Additional documentation == * Tutorial: [[http://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile|Channel file / MRI registration]] * Forum: [[http://neuroimage.usc.edu/forums/showthread.php?1429|Bad MEG-MRI registration]] * Forum: [[http://neuroimage.usc.edu/forums/showthread.php?1389|Coregistration validation]] * Forum: Import the position of SEEG/ECOG contacts: [[http://neuroimage.usc.edu/forums/showthread.php?2206|Post #2206]], [[http://neuroimage.usc.edu/forums/showthread.php?1958|Post #1958]], [[http://neuroimage.usc.edu/forums/showthread.php?2357|Post #2357]] <<EmbedContent(http://neuroimage.usc.edu/bst/get_feedback.php?Tutorials/CoordinateSystems)>> |
Coordinate systems
Author: Francois Tadel
This page describes the various coordinate systems used in Brainstorm, and explains how to convert beetween referentials.
Contents
Subject Coordinate System (SCS / CTF)
Brainstorm uses the CTF head coordinate system. All the surfaces, sensors and additional points are converted into this system when they are imported in Brainstorm.
It is defined in the following way:
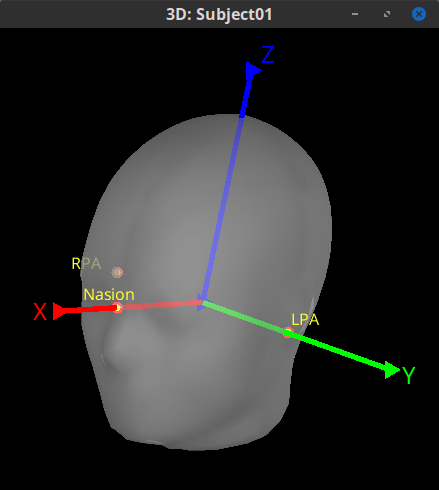
Based on: Nasion, left pre-auricular point (LPA), and right pre-auricular point (RPA).
Origin: Midway on the line joining LPA and RPA.
Axis X: From the origin towards the Nasion (exactly through).
Axis Y: From the origin towards LPA in the plane defined by (NAS,LPA,RPA), and orthogonal. to X axis.
Axiz Z: From the origin towards the top of the head.
The affine transformation from MRI to SCS coordinates is saved in the MRI SCS structure:
SCS.R: [3x3] rotation matrix from MRI coordinates to SCS coordinates.
SCS.T: [3x1] translation matrix from MRI coordinates to SCS coordinates.
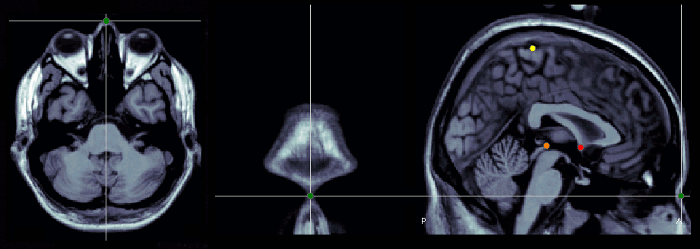
Nasion (NAS)
The nasion is the intersection of the frontal and two nasal bones of the human skull. Its manifestation on the visible surface of the face is a distinctly depressed area directly between the eyes, just superior to the bridge of the nose (source: Wikipedia).
Use the coronal orientation to define it.
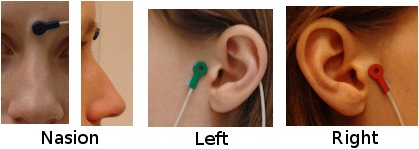
Pre-auricular points (LPA, RPA)
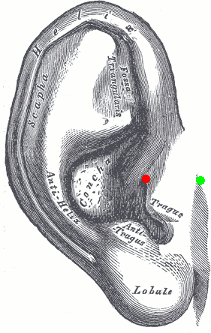
The proper definition of the preauricular point is: "a point of the posterior root of the zygomatic arch lying immediately in front of the upper end of the tragus". This is illustrated with the green point below. The problem with this definition is that it can be really difficult to localize precisely on the MRI. It can lead to severe misregistration between MRI and MEG/EEG. More information on the anatomical location of this point on the FieldTrip website.
Our proposition is to use instead the junction between the tragus and the helix, marked with a red dot in this figure. It can be located much more precisely both anatomically and on the MRI slices.
You can choose to use this definition of the "preauricular point" or not. The most important is to use the same convention for all the steps: digitization of head points (eg. Polhemus) and analysis with Brainstorm.
Use the sagittal orientation to define it.
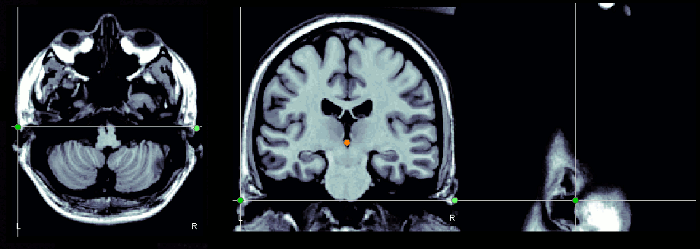
Notes for CTF users
Using a CTF system, you would probably prefer to use the head localization coils to indicate these NAS/LPA/RPA points, instead of these "anatomically correct" points. The coordinate system in the CTF files is based on the position of the three coils you stick on the head of the subject. Typically, the nose coil is slightly above the nasion, and the ear coils about one centimeter more frontal than the points that were previously described.
A good practice is to take pictures of the subject with the coils on, right before bringing him/her in the MEG, and digitize the position of the center of the coils as the nasion/LPA/RPA. Then, when importing the MRI in Brainstorm, try marking as accurately as possible the position of the coils in the MRI. You cannot be very precise at this point, but the errors here are fixed later with the automatic registration.
IMPORTANT: These comments do not apply if you use the Brainstorm Digitize program to drive the Polhemus device. In this case, you are asked to digitize two sets of NAS/LPA/RPA points: the coils and the real anatomical points. Just make a copy of the .pos file saved by Digitize in the .ds folder, then set the real anatomical points in the MRI. Brainstorm is going to automatically handle the transformation between the CTF coils coordinate system and the real subject coordinate system. The result is usually much more accurate and does not require to take pictures of the subjects.
Using a default anatomy
The fiducial points (Nasion, LPA, RPA) used in your recordings might not be the same as the ones used in the anatomy templates in Brainstorm (ICBM152, Colin27, FSAverage). By default, the LPA/RPA points are defined at the junction between the tragus and the helix, as represented with the red dot. If you want to use an anatomy template but you are using a different convention when digitizing the position of these points, you have to modify the default positions of the template with the MRI Viewer.
- Go to the anatomy view
In (default anatomy), right-click on the MRI > Edit MRI
- Modify the position of the fiducial points to match your own convention
- Click on [Save], it will update the surfaces to match the new coordinate system
MRI coordinates
Coordinates system used in the space of the MRI volume, in millimeters:
Axis X: left to right
Axis Y: posterior to anterior
Axis Z: inferior to superior
- The first voxel at the left-posterior-inferior corner of the MRI volume is indexed (1,1,1). The corresponding MRI coordinates of the center of this voxel in millimeters is (voxelsize_x, voxelsize_y, voxelsize_z).
World coordinates
Coordinates system used in the original MRI file, that was loaded as the reference MRI for a subject. It is defined by a 4x4 matrix saved directly in the MRI file. From NIfTI .nii files, it uses the matrix sform if available, or otherwise the matrix qform (see the NIfTI documentation). From FreeSurfer .mgh/.mgz files, it uses the vox2ras matrix (see the FreeSurfer documentation).
In practical terms, it corresponds to the coordinates you see in the title bar when loading your volume in MRIcron, while the coordinates you see in the text boxes correspond in Brainstorm vocabulary to "voxel coordinates". For subject data, it corresponds most of the time to scanner coordinates. For atlases, it may correspond to normalized coordinates (e.g. MNI coordinates).
If available, the transformation from voxels to world coordinates is saved in the field InitTransf{'vox2ras'} of the MRI structure.
MNI coordinates
In order to compare the anatomy of multiple human brains or compute group statistics, it is necessary to define a common reference system. Two types of normalized systems have been widely used in the literature in the past: the atlas defined by Jean Talairach, and the MNI stereotaxic coordinates. You can read about the differences between these two systems here.
While the Talaraich system has been progressively abandonned, the "MNI space" has been increasingly used, thanks to the development of automatic normalization methods in open-source neuroimaging software. Successive updates to the reference brain and software versions led to the coexistence of multiple "MNI spaces". For an exhaustive list, refer to these two websites: BIDS specification, Lead-DBS.
In Brainstorm, MNI coordinates refer to the normalized space defined in SPM12, also referred to as IXI549 space. Like the MRI coordinates above, it is in mm, and the (X, Y, Z) axes are oriented towards (right, anterior, dorsal). The origin is at the AC point (see Talairach section below) of the template, with the (negative) Y axis also going through the PC point of the template.
Computing this MNI normalization for a subject does not alter the input MRI or create a new normalized volume: it only stores a transformation that allows referencing the subject's MRI volume with standardized MNI coordinates. There are two options: a linear and a non-linear approach.
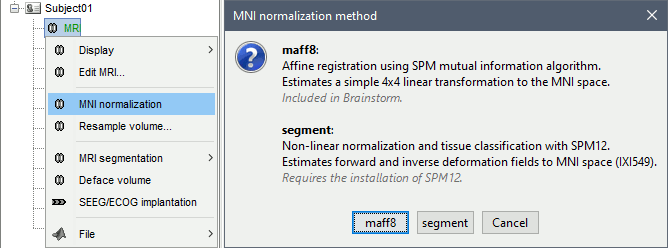
Linear normalization
A [4x4] affine transformation is computed and saved in the MRI NCS structure:
NCS.R: [3x3] rotation matrix from MRI coordinates to MNI coordinates.
NCS.T: [3x1] translation matrix from MRI coordinates to MNI coordinates.
The Brainstorm function mri_normalize_maff.m is based on SPM12 function spm_maff8.m, described in (Ashburner 2005). It is integrated in Brainstorm and does not require the installation of SPM12.
This transformation is fast to compute and to apply to 3D coordinates, and very light in terms of storage. It is appropriate for computing a deformation of the bounding box of the brain and adapted for quickly aligning brains in the same space, or for defining the seed of a volume scout. However, it is not adapted to model the differences in the cortical folding of the individual brains or to the projection of a MNI parcellation to subject space.
Non-linear normalization
Each voxel of the individual brain is projected to the MNI space independently from the others. In order to represent these non-linear deformation fields between volumes, we use the SPM12 representation: three volumes are saved per transformation, one for each dimension (X,Y,Z). One triplet of 3D matrices is necessary to convert from MNI to MRI coordinates (forward transformation), and one other to convert from MRI to MNI (inverse transformation). The following fields are stored in the MRI NCS structure:
NCS.iy: 4D floating point matrix [x,y,z,3]: Inverse MNI deformation field. Same size as the Cube matrix, it gives for each voxel its coordinates in the MNI space, and is therefore used to convert from MRI coordinates to MNI coordinates.
NCS.y: 4D floating point matrix [x,y,z,3]: Forward MNI deformation field. For some MNI coordinates, it gives their coorrespondance in the original subject space. To be interpreted, it has to be used with the matrix y_vox2ras.
NCS.y_vox2ras: [4x4 double], transformation matrix that converts from voxel coordinates of the y volume to MNI coordinates.
NCS.y_method: Algorithm used for the normalization (e.g. 'segment'=SPM12 Segment)
Example: The coordinates of the MRI voxel (i,j,k) correspond in MNI space to:
mni_x = NCS.iy(i,j,k,1); mni_y = NCS.iy(i,j,k,2); mni_z = NCS.iy(i,j,k,3);
A non-linear transformation can represent the individual anatomical variations, and is adapted for warping an MNI parcellation to the subject space. However, it is much heavier computationally. For each MRI volume, we need to store 6 additional volumes in the file, causing an important increase of the file size and the load time. The computation of MNI coordinates is also much longer, slowing down the interface of the MRI Viewer.
Non-linear MNI normalization is available in Brainstorm using the SPM12 Segment algorithm or the CAT12 segmentation pipeline. Both SPM12 and CAT12 can be installed easily as Brainstorm plugins.
References
MNI space: Refers to a system of coordinates for indexing the voxels within a volume, applicable to either individual MRI/CT volumes or anatomical atlases.
Anatomy template: Set of anatomical files (MRI, surfaces, parcellations) representing a brain atlas. As the reference anatomy, we typically use the ICBM125 2009c Nonlinear Asymmetric, but other options are described in the tutorial Using the anatomy templates. An anatomy template can be used for various purposes:
- Replacing the subject anatomy in the Brainstorm database when no individual scans are available for the subject. Setting the anatomy of the subject is necessary for 2D/3D topography display and source estimation.
Warping an anatomy template to match the digitized head shape of a subject. See tutorial: Warping the anatomy templates
For group analysis at the source level: an intermediate step of projection on an anatomy template is necessary. See tutorial: Group analysis: Subjects coregistration
MNI template: Anatomy template that has the particularity to be registered into an MNI space (MNI152, Colin27).
MNI parcellations: Volume of integers where each value represents an anatomical label, and registered to an MNI space. A non-linear deformation field can be applied to an MNI parcellation in order to create a volume parcellation in subject space. The anatomical regions can be used as volume scouts, for reference in the MRI viewer, or for labelling SEEG contacts. See also tutorial Using the anatomy templates.
MacOS troubleshooting
Error "mexmaci64 cannot be opened because the developer cannot be verified":
ACPC coordinates
This coordinate system is recommended for iEEG coordinates in the BIDS specification. It relies on two points, the anterior and posterior commissures (AC and PC), and the midsagittal plane. The third point needed here, the interhemispheric point (IH), is used to define this plane. The anatomical landmarks are determined in the individual's anatomical scan and no scaling or deformations have been applied to the individual's anatomical scan.
Origin: Anterior commissure (AC)
Axis X: From left to right
Axis Y: Passing through PC and AC, from posterior to anterior
Axis Z: Along the mid-saggital plane (defined from AC, PC and IH), from inferior to superior
References:
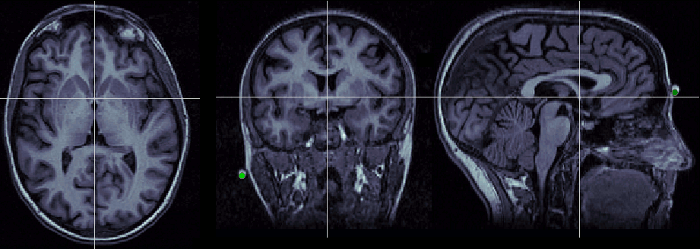
Anterior commissure (AC)
Description of the anterior commissure at this Wikipedia page.
Technique to localize it:
- Use the axial orientation.
- Start from a slice just below the corpus callosum and move around it (slightly up or down)
- You are looking for a small fiber tract that connects the two hemispheres
- You should find two spots corresponding to this description. The more frontal one is the anterior commissure, the other one is the posterior commissure
- Once you have localized it: if it is visible on more than one slice, use the upper one (the one closest to the top of the head).
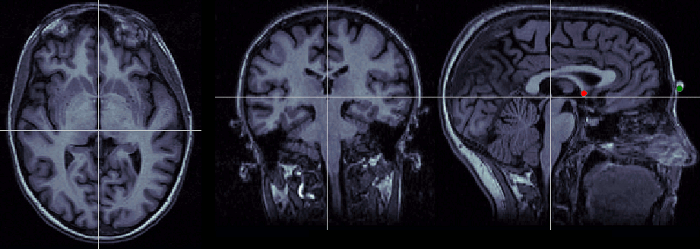
Posterior commissure (PC)
Description of the posterior commissure at this Wikipedia page.
Technique to localize it:
- Use the axial orientation
- It is a fiber tract similar to the anterior commissure, but a few centimeters more posterior.
- If it is visible on more than one slice, pick the lowest one.
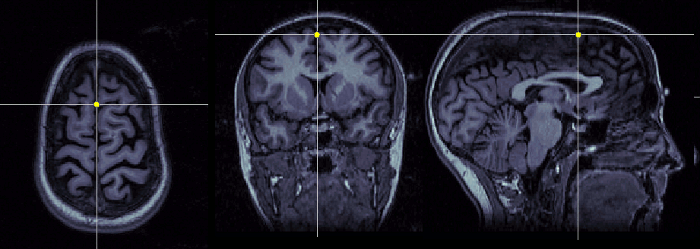
Interhemispheric point (IH)
Pick any point in the interhemispheric space, somewhere in the top of the head. Do not use a point too close from the commissures.
Talairach coordinates
Similar to the ACPC coordinates, but with an additional piecewise scaling to match the Talairach-Tournoux (1988) atlas. See FieldTrip documentation. No used in Brainstorm.
CapTrak coordinates
This coordinate system is accepted for EEG coordinates in the BIDS specification. It relies on the three head points LPA/RPA/NAS described above.
Origin: Between LPA and RPA
Axis X: From LPA through RPA exactly
Axis Y: Orthogonal to the X-axis through the nasion (NAS)
Axis Z: Orthogonal to the XY-plane through the vertex of the head
References:
Neuromag coordinates
This coordinates system is not used in Brainstorm, and described here only FYI. All the information contained in Neuromag .FIF files are in this CS, and converted to Brainstorm SCS at importation.
It is defined in the following way:
Based on: Nasion, left pre-auricular point (LPA), and right pre-auricular point (RPA).
Origin: Intersection of the line L through LPA and RPA and a plane orthogonal to L and passing through the nasion.
Axis X: From the origin towards the RPA point (exactly through)
Axis Y: From the origin towards the nasion (exactly through)
Axiz Z: From the origin towards the top of the head
Converting between coordinate systems
The conversions between coordinate systems is handled with one single function cs_convert:
sMri: MRI structure from the database: right-click on the MRI file > File > Export to Matlab > sMri
- src/dest: Source and destination coordinates systems: {'voxel', 'mri', 'scs', 'mni', 'world'}
- P: List of points [Npoints x 3]
All the the coordinates have to be in meters (not millimeters).
Pdest = cs_convert(sMri, 'src', 'dest', Psrc);
Examples:
P_mri = cs_convert(sMri, 'voxel', 'mri', P_voxel); % Voxel => MRI coordinates P_mni = cs_convert(sMri, 'scs', 'mni', P_scs); % SCS => MNI coordinates
Brainstorm .pos files
The .pos files generated with the Brainstorm Digitizer contain both the positions of the anatomical landmarks (nasion, left ear, right ear) and the position of the head localization coils (HPI).
Having the two sets of points allows us to convert automatically from the native CTF coordinates (based on the coils) to the recommended landmarks in Brainstorm (based on the anatomical landmarks). When a Brainstorm .pos file is present in the CTF .ds folder, it is loaded automatically and used to convert the positions of the EEG and MEG sensors to the anatomical reference. This is done automatically without any message or user confirmation. In this case, you should mark the anatomical landmarks (real nasion and tragus/helix junctions) in the MRI Viewer when importing the anatomy. This is what is described in the introduction tutorials.
Otherwise, if you are importing CTF recordings without a Brainstorm .pos file in the .ds folders, or if you are importing the .pos files after linking the recordings to the database, you should typically select the positions of the CTF HPI coils in the MRI viewer.
The syntax of these .pos files is the following, one line per digitized point:
Number of EEG electrodes Index Label X Y Z : Defines an EEG electrode Index X Y Z : Defines a head shape point Label X Y Z : Defines a reference point
In the .pos files, the anatomical landmarks are labelled Nasion/LPA/RPA, and the CTF HPI coils are labelled HPI-N/HPI-L/HPI-R (for nasion coil, left coil, right coil). Note that in the CTF software, the HPI coils are labelled NAS/LPA/RPA, which can lead to some confusion.
We usually take multiple measures of each reference point and average them when we load the file. This helps improving the accuracy of the registration.
2 1 Cz 5.29886357 -0.39211620 13.97175211 2 Pz -2.72999896 0.56457819 14.80592908 3 9.47286768 0.09984190 -2.22056385 4 9.61745050 -0.00705346 -2.64471836 [...] 242 2.68917264 3.02778502 13.88281027 243 1.10525214 3.54043941 14.17347208 Nasion 9.59228188 0.08328658 -1.88504105 LPA -0.47469294 6.81987024 0.62870235 RPA -0.96029260 -6.72333418 0.16475241 Nasion 9.76823213 -0.11917776 -1.87417223 LPA -0.29274026 6.88415084 0.70120923 RPA -0.99183277 -6.70128616 0.08443862 HPI-N 10.61990713 0.01532629 0.00407281 HPI-L 0.20165313 6.74350937 -0.00134360 HPI-R -0.23124566 -6.76723759 -0.00008921 HPI-N 10.61095674 -0.01532629 -0.00407281 HPI-L 0.27017080 6.81335558 0.00134360 HPI-R -0.24057827 -6.78962736 0.00008921
Additional documentation
Tutorial: Channel file / MRI registration
Forum: Bad MEG-MRI registration
Forum: Coregistration validation
Forum: Import the position of SEEG/ECOG contacts: Post #2206, Post #1958, Post #2357
