What's next
A roadmap to the future developments of Brainstorm.
Contents
Recordings
- Sleep scoring wish list (Emily C):
- Configurable horizontal lines (for helping detecting visually some thresholds)
- Mouse ruler: Measure duration and amplitude by dragging the mouse.
- Add filter option to the montage editor: This would allow splitting the signals in various frequency bands.
- Automatic spindle detector
https://neuroimage.usc.edu/forums/t/page-overlap-while-reviewing-raw-file-a-way-to-set-to-0/11229/13
- Montage:
- Allow saving ICA results as new file using process "Apply montage"
- RAW file viewer:
- Downsample before filtering? (attention to the filter design)
- Add parameter to make the visual downsampling more or less aggressive
- Pre-load next page of recordings
- Keep the filter specifications in memory instead of recomputing for every page
- Bad trials: When changing the status of bad to good: remove the bad segments as well, otherwise it is not processed by processes like the PSD.
- Review clinical recordings: Reduce the dimensionality of the data with a simple inverse problem, similar to what we do for the magnetic extrapolation ("Regional sources" in BESA, cf S Rampp)
- MEG/EEG registration: Apply the same transformation to multiple runs
- Create heat maps: Maybe with matlab function heatmap?
Interface
Add a warning when computing a forward model with > 100000 sources (check selection)
- Snapshot: Save as image / all figures (similar to Movie/all figure)
Generalize the use of the units (field .DisplayUnits): Rewrite processes to save the units correctly
- Colormaps:
- Allow brightness/contrast manipulations on the custom colormaps
- Global colormap max: Should get the maximum across all the open files
- Copy figures to clipboard (with the screencapture function)
- Smooth display from figure_image (ERPimage, raster plot...)
Contact sheets & movies: use average of time windows instead of single instants, for each picture.
- Contact sheets: Allow explicit list of times in input (+ display as in MNE-Python with TS)
- Display CTF coils: Show discs instead of squares
- Use boundary() instead of conhull() in all the display functions (ie. 2DDisc)
- Progress bar: Add a "Cancel" button
- Error message: Add a link to report directly the bug on the forum
Reorganize menus (Dannie's suggestion):
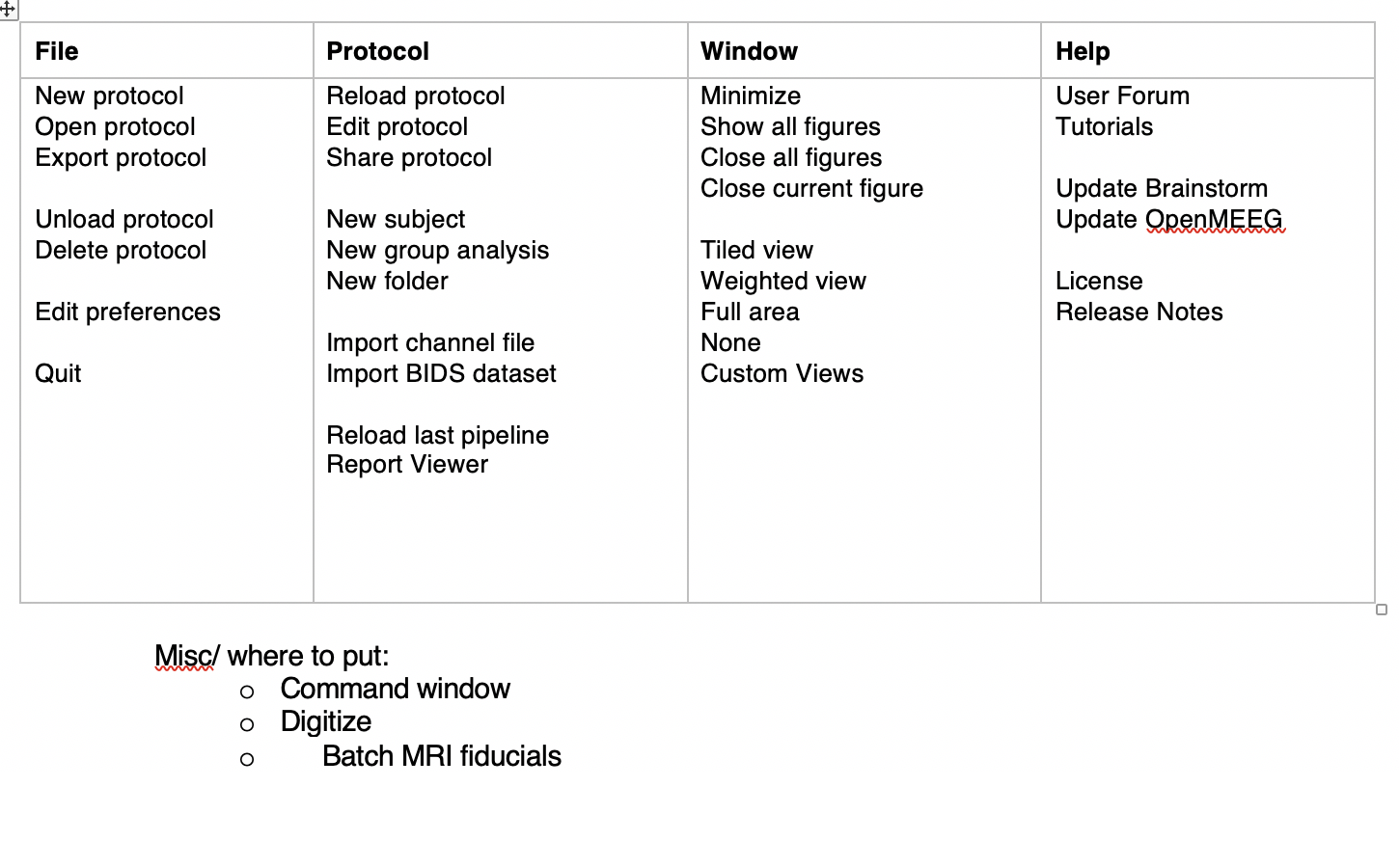
Connectivity
- Thresholding and stat tests the connectivity matrices
- Connectivity on unconstrained sources: "Default signal extraction for volume grids should be the time series of the first principal component of the triplet signals after each has been zero-meaned" (SB)
- Display of connectivity graphs:
- Display as straight lines
- Recode 2D graphs
- 3D display with anatomical constrains
- Display using real position of EEG electrodes
- Use new band-pass filters in bst_connectivity ('bst-hfilter' instead of 'bst-fft-fir')
- Matrix view of NxN graphs: Add legend of the elements along X and Y axis
- Graph view:
- Does not display negative values correctly (correlation or difference of coherence)
- Re-write using pure Matlab code and smoothed graphics
- Fixed scales for intensity sliders
- Text bigger
- Too much data in appdata
- Fixed scales for intensity sliders
- Add "=" shortcut for having graphs with similar configurations
- Disable zoom in one region (serious bugs)
- NxN on sensors: does not place the sensors correctly in space
- Coherence:
Average cross-spectra instead of concatenating epochs (to avoid discontinuities)
Explore inter-trial approaches (Esther refers to chronux toolbox)
- Granger: Check for minimum time window (Esther: min around 500-1000 data points)
- PLV:
- Add p-values
- Remove evoked
- Optimize code
- Add time integration
- Unconstrained sources
- Add warning when running of short windows (because of filters)
- Time-resolved correlation/coherence: Display as time bands
Tutorial coherence [1xN] : Reproduce FieldTrip results?
Connect NxN: Display as time series > Display warning before trying to open too many signals
Processes
- Decoding/Classifiers: Implement Dimitrios and scikit-learn algorithms
- Allow processes in Python and Java
- Add MNE-Python functions:
- scikit-learn classifiers
- Implement data exchange with MNE-Python: write FIF files from Brainstorm and/or pass python objects in memory instead of FIF files
https://neuroimage.usc.edu/forums/t/best-way-to-export-to-mne-python/12704/3
- SSS/tSSS cleaning
- Reproduce other tutorials / examples
- Change the graphic renderer from Matlab
Add FieldTrip functions:
- ft_sourceanalysis:
- Check noise covariance
- Check all the options of all the methods
- Single trial reconstructions + noise covariance?
Filters?? http://www.fieldtriptoolbox.org/example/common_filters_in_beamforming
Beamformers: Save ftSource.avg.mom
http://www.fieldtriptoolbox.org/workshop/meg-uk-2015/fieldtrip-beamformer-demohttp://www.fieldtriptoolbox.org/tutorial/beamformingextended
- Baseline? Two inputs?
ft_prepare_sourcemodel: Compute MNI transformation (linear and non-linear) => Peter
- ft_prepare_heamodel: Add support from BEM surfaces from the Brainstorm database
- Freqanalysis: ITC
- ft_read_atlas('TTatlas+tlrc.BRICK');
ft_volumereslice: http://www.fieldtriptoolbox.org/faq/how_change_mri_orientation_size_fov
- ft_freqanalysis
- ft_combineplanar
- ft_sourceanalysis:
- Optimization:
- Use CUDA for speeding up some operations (filtering, wavelets, etc)
- Use Matlab Coder to optimize: Wavelets, bandpass filter, sinusoid removal
- Pipeline editor:
- Bug: After "convert to continuous", the time of the following processes should change
- Add loops over subjects/conditions/trial groups
- Events: Allow selection from a drop-down list (similar to option "channelname" in panel_process_selection)
- When computing sources from the pipeline editor: doesn't reselect the options if you click twice on "edit" (works for minnorm, but not for lcmv)
ITC: Inter-trial coherence (see MNE reports for group tutorial)
http://www.sciencedirect.com/science/article/pii/S1053811916304232- ICA:
- Why doesn't the ICA process converge when using 25 components in the EEG tutorial?
- Add an option to resample the signals before computing the ICA decomposition
- Exploration: Add window with spectral decomposition (useful for muscle artifacts)
- Export IC time series (and then compute their spectrum): solves the problem above
Comparison JADE/Infomax:
http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0030135- Use faster methods (MNE-Python?)
- Add methods: SOBI, Fastica, AMICA/CUDICA (recommended by S Makeig)
Dimension reduction with PCA adds artifacts: Not done by default in EEGLAB
Contact: Stephen Shall Jones ( shall-jones@infoscience.otago.ac.nz )
Student Carl Leichter detailed this in his thesis- S Makeig: Use ICA to select the IC of interest instead of only removing artifacts
- Display of spectrum for components (PSD/FFT)
- Import ICA matrices available in EEGLAB .set files
EEGLAB recommends ICA + trial rejection + ICA again: Impossible right now with Brainstorm
(http://sccn.ucsd.edu/wiki/Chapter_09:_Decomposing_Data_Using_ICA)ICA+machine learning: https://www.ncbi.nlm.nih.gov/pubmed/28497769
Automated artifact rejection: https://arxiv.org/abs/1612.08194
- Save IC time series in database
Use EYE-EEG: EEGLAB toolbox for eye-tracker guided ICA (Olaf Dimigen): http://www2.hu-berlin.de/eyetracking-eeg/
- Other EEGLAB functions:
Step function detection: https://github.com/lucklab/erplab/wiki/Artifact-Detection:-Tutorial
- SSP:
Display warning if changing the ChannelFlag while there is a Projector applied
- Spectral flattening (John):
- ARIMA(5,0,1): Apply on the signal before any frequency/connectivity/PAC analysis
- PSD:
- Rewrite to have the same input as coherence (frequency resolution instead of window length)
Allow display of Avg+StdErr
Remove line noise: http://www.nitrc.org/projects/cleanline
- Time-frequency:
- Optimization: bst_timefreq (around l.136), remove evoked in source space: Average should be computed in sensor space instead of source space (requested by Dimitrios)
Short-time Fourier transform: http://www.mikexcohen.com/lectures.html
Matching pursuit: http://m.jneurosci.org/content/36/12/3399.abstract?etoc
- Bug: Display logs as negative
- Bug: 3D figures: Colormaps with "log" option doesn't work
- Bug: Difference of power displayed in log: problems (Soheila)
- 2D Layout in spectrum
- Make much faster and more memory efficient (C functions coded by Matti ?)
- TF scouts: should display average of TF maps
- Impossible to keep complex values for unconstrained sources
- Pad short epochs with zero values for getting lower frequencies
- Hilbert with time bands very slow on very long files (eg. 3600s at 1000Hz) because the time vector is still full (10^7 values): save compressed time vector instead.
- Extend clusters tab to display of TF to overlay TF signals (Svet)
- When normalizing with baseline: Propagate with the edge effects marked in TFmask
- Allow baseline normalization of files computed with time bands
- Allow running TF on montages
- Review continuous files in time-frequency space (for epilepsy)
- Bug when computing TF on constrained and unconstrained scouts at the same time (in mixed head models for instance): uses only the constrained information and doesn't sum the 3 orientations for the unconstrained regions.
- Artifact detection:
- Artifact rejection like SPM: if bad in 20%, bad everywhere
- Test difference between adjacent samples
- Events detection: Add option "std" vs "amplitude"
- Simulation:
EEGSourceSim: https://www.sciencedirect.com/science/article/pii/S0165027019302341
Fix units in simulation processes => no *1e-9 in "simulate recordings"
- Use "add noise" process from Hui-Ling (in Work/Dev/Divers)
- Use field process field "Group" to separate Input/Processing/Output options
- Use new Matlab functions: movmean, movsum, movmedian, movmax, movmin, movvar, movstd
Database
- Faster DB searches (for Emily)
- Add buttons to sort files: by name, by comment, by date
- MEG protocols: More flexible organization of the database; sub-conditions to allow different runs X different conditions.
- Matrix files: Allow to be dependent from other files
- Rename multiple files
- Default headmodel lost when reloaded: Keep selection on the hard drive (in brainstormstudy.mat)
- Auto-save:
protocol.mat can be too big: do not store the results links in it (and recreate when loading)- http://neuroimage.usc.edu/forums/t/abnormally-slow-behavior/2065/10
- Improve auto-save: add tracking file next to protocol.mat, do not save all the time, only when closing app, and reload protocol at stratup if tracking file is still there
Distributed computing
Options from FieldTrip:
Loose collection of computers: https://github.com/fieldtrip/fieldtrip/tree/master/peer
Alternative, with less limitations: http://research.cs.wisc.edu/htcondor/
Single multicore machine: https://github.com/fieldtrip/fieldtrip/tree/master/engine
Batch system: https://github.com/fieldtrip/fieldtrip/tree/master/qsub
Documentation: http://fieldtrip.fcdonders.nl/faq#distributed_computing_with_fieldtrip_and_matlab
Various initiatives: http://samirdas.github.io/Data_sharing.html#/
Source modeling
Use eLORETA instead of sLORETA?
https://neuroimage.usc.edu/forums/t/compute-eeg-sources-with-sloreta/13425/6"eLORETA algorithm is available in the MEG/EEG Toolbox of Hamburg (METH)": https://www.biorxiv.org/content/biorxiv/early/2019/10/17/809285.full.pdf
- Point-spread and cross-talk functions (code in MNE-Python):
- Dipoles:
- Project individual dipoles files on a template
- panel_dipoles: Doesn't work with multiple figures
- Project sources: Very poor algorithm to project sub-cortical regions and cerebellum (algorithm to fit surfaces should be imrpoved)
Menu head model > Copy to other conditions/subjects (check if applicable first)
Menu Sources > Maximum value: Doesn't work with volume or mixed head models
- Mixed head models:
- Set loose parameter from the interface
- Volume grid:
- Optimize: 3D display (better than 9x9 cubes)
- Optimize: vol_dilate (with 26 neighbors)
Menu Sources > Simulate recordings:
- Do not close the 3D figures after generating a new file
- Add a process equivalent to this menu
- Panel Get coordinates: Add button "find maximum"
- BEM single sphere: Get implementation from MNE
- Unconstrained sources:
- Stat and connectivity: what to do? (re-send email John+Sylvain)
Sources on surface: Display peak regions over time (time = color) => A.Gramfort
- Process "Extract scouts time series": Add PCA option (replace isnorm with choice PCA/Norm)
- Add eyes models to attract eye activity
Display source maps on a flat 2D cortex projection (Mollweide projection): https://neuroimage.usc.edu/forums/t/source-model-display-and-output/13940/5
Anatomy
- Multi-Scale Brain Parcellator (Lausanne2008):
- MNI transformation: Use SPM non-linear MNI transformation y_...
- Registration:
Getting electrode positions from 3D scanners: https://sccn.ucsd.edu/wiki/Get_chanlocs
Use the same registration for multiple recording sessions that have already re-registered previously (eg. with MaxFilter)
- When linking multiple EEG recordings including 3D positions, do the registration only once and copy it to all the runs
- Compute non-linear MNI registration instead of linear
- Select and remove bad digitized head points before automatic coregistration
Load the MNE -transf.fif: http://neuroimage.usc.edu/forums/showthread.php?2830
- MRI Viewer:
- Pan in zoomed view (shift + click + move?)
- Zoom in/out with mouse (shift + scroll?)
- Ruler tool to measure distances
- Display scouts as additional volumes
Render surface envelope in the MRI as a thin line instead of the full interpolation matrix
Or use inpolyhedron to get a surface mask and then erode it to get the volume envelope- Edit fiducials: Replace 6 text boxes with 1 for easy copy-paste (see fiducials.m)
Optimize computation interpolation MRI-surface (tess_tri_interp) => spm_mesh_to_grid
- CAT12: Optimize interpolation of atlases (import is super slow...)
BrainSuite:
Add new labels to all BrainSuite anatomy templates
- Use same colors for left and right for anatomical atlases
Pediatric head atlases: https://www.pedeheadmod.net/pediatric-head-atlases-v1-2/
- Scouts:
- Display edges in the middle of the faces instead of the vertices
- Display scouts in a tree: hemisphere, region, subregion
- Sort scouts by region in process options
- Downsample to atlas: allow on timefreq/connect files
Project from one hemisphere to the other using registered spheres/squares (http://neuroimage.usc.edu/forums/t/how-to-create-mirror-roi-in-the-other-hemisphere/5910/8)
- Parcellating volume grids: scikit-learn.cluster.Ward
- Major bug when importing surfaces for an MRI that was re-oriented manually
Surface>Volume interpolation: Use spm_mesh_to_grid
- Bug: Hide scouts in the preview of the grid for volume head models
Geodesic distance calculations:
https://www.mathworks.com/matlabcentral/fileexchange/6110-toolbox-fast-marching
ECOG/SEEG
- Contact positions: Import / set / detect
- New option: Align on none|inner|cortex to replace ECOG-mid
- Add history: Save modifications and transformations applied to the channel files (Marcel)
- Project contact positions across subjects or templates (Marcel)
- Add menu to import implantation channel file in imported recordings
- Automatic segmentation of CT:
- ECOG:
- Project and display contacts on cortex surface should consider the rigidity of the grids: Contacts cannot rotate, and distance between contacts should remain constant across runs
Method for contacts projection: https://pdfs.semanticscholar.org/f10d/6b899d851f3c4b115404298d7b997cf1d5ab.pdf
ECOG: Brain shift: When creating contact positions on a post-implantation image, the brain shift should be taken into account for creating images of the ECOG contacts on the pre-op brain => iELVis (http://ielvis.pbworks.com/w/page/116347253/FrontPage)
- Display:
- Bad channels: Contacts greyed out instead of ignored (Marcel)
- Display time in H:M:S
- Display curved SEEG electrodes
Export list of contacts with a probability of anatomical regions from various atlases: https://neuroimage.usc.edu/forums/t/seeg-contacts-anatomical-location/14756
Detection CEEP stim artifacts: Use ImaGIN code ImaGIN_StimDetect
Statistics
- ANOVA:
- Which functions to use?
- Write panel similar to Process1 and Process2 to allow the
- Output = 1 file per effect, all grouped in a node "ANOVA"
- Display several ANOVA maps (from several files) on one single figure, using a "graphic accumulator", towards which one can send any type of graphic object
Quality control before statistics, on condition averages across subjects:
mean(baseline)/std(baseline): shows bad subject quickly.Use SurfStat: Impements interesting things, like an analytical cluster-based p-value correction (Random-field theory which is used in SPM) - Peter
- Export to R or SPSS for advanced stat
Input / output
XDF import: Use FieldTRip or the EEGLAB plugin, contact Martin Bleichner (Oldenburg)
https://github.com/sccn/xdf/blob/master/xdf_sample.xdf- DICOM converter:
- Add dcm2nii (MRICron)
- Add MRIConvert
FieldTrip: Import/Export time-frequency:
- 4D file format:
- Use reader from MNE-Python: mne.io.read_raw_kit (doesn't require Yokogawa slow library)
- Reference gradiometers: Keep the orientation of the first or second coil?
- Reference gradiometers: Add the sensor definition from coil_def.dat
- Validate with phantom recordings that noise compensation is properly taken into account
- The noise compensation is considered to be always applied on the recordings, not sure this assumption is always correct
- 4D phantom tutorial (JM Badier?)
- EEG File formats:
EEG CeeGraph
- EGI: Finish support for epoched files (formats 3,5,7)
Persyst .lay: https://github.com/ieeg-portal/Persyst-Reader
Nervus .eeg: https://github.com/ieeg-portal/Nervus-Reader
Biopac .acq: https://github.com/ieeg-portal/Biopac-Reader
- gTec EEG recordings: Read directly from the HDF5 files instead of the Matlab exports.
- BCI2000 Input (via EEGLAB plugin)
- BST-BIN: Add compression to .bst
- Review raw on all the file formats (ASCII EEG and Cartool missing)
- SPM .mat/.dat: Fix the import of the EEG/SEEG coordinates
- Get acquisition date from files: Missing for 4D
Distribution & documentation
- Tutorial OMEGA/BIDS:
- Add review of literature for the resting state MEG
- Download example datasets directly from the OMEGA repository
New tutorials:
Other public datasets: https://github.com/INCF/BIDS-examples/tree/bep008_meg/
- Rat PAC + high gamma (Soheila)
- EEG/research
FieldTrip ECOG tutorial: http://www.fieldtriptoolbox.org/tutorial/human_ecog
FieldTrip cortico-muscular coherence tutorial: http://www.fieldtriptoolbox.org/tutorial/coherence
Reproduce tutorials from MNE-Python: https://martinos.org/mne/stable/tutorials.html
Cam-CAN database: https://camcan-archive.mrc-cbu.cam.ac.uk/dataaccess/<<BR>>(download new datasets, including maxfiltered files and manual fiducial placements)
- MEG steady-state / high-gamma visual / frequency tagging
- BIDS-EEG example datasets
- Stand-alone ICA tutorial
- Move all the files to download to the cloud for faster download everywhere in the world
- Provide secure way of sending password over HTTPS for:
- Account creation
- Forum exchanges
org.brainstorm.dialog.CloneControl
Workflows FieldTrip: http://www.fieldtriptoolbox.org/faq/what_types_of_datasets_and_their_respective_analyses_are_used_on_fieldtrip
Count GitHub clones in the the download stats
- Deface the MRIs of all the tutorials
- Clean up the wiki:
- Remove all the wiki pages that are not used
- Check all the links in all the pages
- Check that all the TODO blocks have been properly handled
- Remove useless images from all tutorials
- Update page count on the main tutorials page
Current bugs
- Image viewer:
- Difficult to get to 100%
- Buggy on some systems
- 2DLayout:
- (TF) Units are weird with % values
- (TF) Difficult to navigate in frequencies: Scaling+changing frequency resets the scaling
- Progress bar:
- Doesn't close properly on some Linux systems
- Focus requests change workspace when processing constantly (Linux systems)
- MacOS bugs:
- Buttons {Yes,No,Cancel} listed backwards
- Record tab: Text of epoch number is too big
- Colormap menus: Do not work well on compiled MacOSX 10.9.5 and 10.10
- in_bst_data_multi: If trials have different sizes, output is random (the one of the first file)
- Canolty maps computation: Fix progress bar
Geeky programming details
- bst_bsxfun: After 2016b, we can use directly the scalar operators (./ .* ...) instead of bsxfun. Update bst_bsxfun to skip the use of bsxfun when possible.
Interface scaling: Rewrite class IconLoader to scale only once the icons at startup instead of at each request of an icon (might improve the speed of the rendering of the tree)
- Hide Java panels instead of deleting them
- Processes with "radio" and "radio_line" options: Replace with "radio_label" and "radio_linelabel"
- Interpolations: Use scatteredInterpolant, griddedInterpolant, triangulation.nearestNeighbor (2014b)
- bst_warp and channel_project: Use tess_parametrize_new instead of tess_parametrize
- Shared kernels: "get bad channels" operation in a different way (reading all the files is too slow)
- Optimize bst_get:
- Now study and subject have necessarily the same folder name
- Replace big switch with separate functions
- Fix all the 'todo' blocks in the code
