|
Size: 5830
Comment:
|
Size: 10456
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 4: | Line 4: |
| Practical workshop on EEG/MEG analysis at ICM, in parallel with FieldTrip and MNE-Python.<<BR>>[[http://practicalmeeg2019.org/|Workshop website]] | Practical workshop on EEG/MEG analysis at ICM, in parallel with FieldTrip and MNE-Python. Program available on the workshop website: http://practicalmeeg2019.org/ |
| Line 6: | Line 6: |
| This session is based on a simplified version of the [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity|SEEG/Epileptogenicity tutorial]]. | The hands-on part of this workshop is based on the Frontiers Research Topic [[https://www.frontiersin.org/research-topics/5158/from-raw-megeeg-to-publication-how-to-perform-megeeg-group-analysis-with-free-academic-software|From raw MEG/EEG to publication]]: a collection of articles processing the [[https://openneuro.org/datasets/ds000117|same multimodal dataset]] using different software environments. We will process together one subject of this dataset, following the processing pipeline described in the article [[https://www.frontiersin.org/articles/10.3389/fnins.2019.00076/full|MEG/EEG Group Analysis With Brainstorm]] and the corresponding [[https://neuroimage.usc.edu/brainstorm/Tutorials/VisualSingle|online tutorial]]. |
| Line 11: | Line 11: |
| <<HTML(<TR><TD>)>>'''Where'''<<HTML(</TD><TD>)>>Telecom ParisTech<<HTML(</TD></TR>)>> | <<HTML(<TR><TD>)>>'''Where'''<<HTML(</TD><TD>)>>ICM<<HTML(</TD></TR>)>> |
| Line 13: | Line 13: |
| <<HTML(<TR><TD>)>>'''When'''<<HTML(</TD><TD>)>>Monday July 2nd, 2018: 8:30-12:15<<HTML(</TD></TR>)>> | <<HTML(<TR><TD>)>>'''When'''<<HTML(</TD><TD>)>>Tuesday Dec 3rd - Thursday Dec 5, 2019: 8:30-17:30<<HTML(</TD></TR>)>> |
| Line 15: | Line 15: |
| <<HTML(<TR><TD>)>>'''Instructors'''<<HTML(</TD><TD>)>>[[AboutUs/FrancoisTadel|Francois Tadel]] (Grenoble Institute of Neuroscience)<<BR>>[[http://www.neurotrack.fr/contact/|Anne-Sophie Dubarry]] (Aix-Marseille University)<<HTML(</TD></TR>)>> | <<HTML(<TR><TD>)>>'''Instructors'''<<HTML(</TD><TD>)>>[[AboutUs/FrancoisTadel|Francois Tadel]] (Grenoble Institute of Neuroscience)<<BR>>[[http://www.neurotrack.fr/contact/|Anne-Sophie Dubarry]] (Aix-Marseille University)<<BR>>[[https://cognition.ens.fr/fr/member/8648/aurelien-weiss|Aurélien Weiss]] (ENS)<<HTML(</TD></TR>)>> |
| Line 19: | Line 19: |
| <<HTML(<TR><TD>)>>'''Slides'''<<HTML(</TD><TD>)>>[[https://neuroimage.usc.edu/resources/cuttingeeg2018_brainstorm_slides.pdf|Introduction]] <<HTML(</TD></TR>)>> | <<HTML(<TR><TD>)>>'''Documents'''<<HTML(</TD><TD>)>>[[https://neuroimage.usc.edu/resources/practicalmeeg2019_brainstorm.pdf|Introduction slides]] | [[http://neuroimage.usc.edu/resources/practicalmeeg2019_brainstorm_walkthrough.pdf|Hands-on walkthrough]] | [[https://github.com/brainstorm-tools/brainstorm3/blob/master/toolbox/script/tutorial_practicalmeeg.m|Processing script]] | [[http://neuroimage.usc.edu/resources/practicalmeeg2019_anatomy_soft.pdf|Anatomy processing]]<<HTML(</TD></TR>)>> |
| Line 24: | Line 24: |
| The participants are required to <<HTML(<FONT color=red>)>> '''bring a laptop''' <<HTML(</FONT>)>> (an external mouse will add to your comfort). In order to make the session as efficient as possible, we ask all the attendees to <<HTML(<FONT color=red>)>>'''download, install and test'''<<HTML(</FONT>)>> the software and sample dataset on their laptops prior to the workshop. | The participants are required to <<HTML(<FONT color=red>)>> '''bring a laptop''' <<HTML(</FONT>)>> (an external mouse will add to your comfort). In order to make the session as efficient as possible, we ask all the attendees to <<HTML(<FONT color=red>)>>'''download, install and test'''<<HTML(</FONT>)>> the software and sample dataset on their laptops prior to the workshop. |
| Line 26: | Line 26: |
| Please read carefully the following instructions: <<BR>>[[WorkshopTalca2018Install|How to prepare your laptop for the training]] | Please read carefully the installation instructions: <<BR>>[[WorkshopParis2019Install|How to prepare your laptop for the training]] |
| Line 28: | Line 28: |
| == Program: Day 1 == <<HTML(<TABLE>)>> |
== Program == === 1. Tuesday am: From raw to ERP === * Introduction to Brainstorm (lecture) * Review the MEG+EEG recordings * Read and organize events * Frequency filters * Artifacts detection and cleaning |
| Line 31: | Line 36: |
| <<HTML(<TR><TD>)>>8:30-9:00<<HTML(</TD><TD>)>>Onsite assistance in installing the material for the training session <<HTML(</TD></TR>)>> | === 2. Tuesday pm: Sensor level analysis === * Epoching and averaging * Time-frequency analysis |
| Line 33: | Line 40: |
| <<HTML(<TR><TD>)>>9:00-10:00<<HTML(</TD><TD>)>>'''Lecture: Brainstorm overview (Tadel''') | === 3. Wednesday am: Creating head and source models === * Import anatomy * Registration MRI-sensors * Forward model * Simulations |
| Line 35: | Line 46: |
| Software structure, typical data workflow <<HTML(</TD></TR>)>> | === 4. Wednesday pm: Single and distributed sources === * MEG noise covariance: From empty room recordings * EEG noise covariance: From pre-stimulus baselines * Distributed sources / minimum norm estimation * Regions of interest * LCMV Beamformer * Dipole scanning / Volume source models |
| Line 37: | Line 54: |
| <<HTML(<TR><TD>)>>10:00-10:45<<HTML(</TD><TD>)>>'''Practice''' | === 5. Thursday am: Group-level analysis === * Project source maps on MNI template * Statistical testing * Export data to other programs (FieldTrip, SPM, ...) * Scripting * Example for this workshop: https://github.com/brainstorm-tools/brainstorm3/blob/master/toolbox/script/tutorial_practicalmeeg.m |
| Line 39: | Line 61: |
| Database explorer | === 6. Thursday pm: User requests === * Bring your own data or prepare your questions |
| Line 41: | Line 64: |
| MRI volumes, surfaces and anatomical atlases<<HTML(</TD></TR>)>> | == Installation instructions == Before coming to the workshop, you need to download the software and the tutorial dataset (1.6 Gb). To streamline troubleshooting during the session, please save all the downloaded files '''__on your Desktop__'''. |
| Line 43: | Line 67: |
| <<HTML(<TR><TD>)>>10:45-11:00<<HTML(</TD><TD>)>>''Coffee break''<<HTML(</TD></TR>)>> | 1. From the Brainstorm [[http://neuroimage.usc.edu/bst/download.php|Download]] page, log in or create a Brainstorm account 1. Download <<HTML(<FONT color=red>)>>brainstorm_YYMMDD.zip<<HTML(</FONT>)>> (75 Mb) 1. If you already have Brainstorm: update it. We need a version released '''after November 18th''' 1. Download the tutorial dataset (1.5 Gb):<<BR>>https://owncloud.icm-institute.org/index.php/s/cNu5jmiOhe7Yuoz/download 1. Unzip the two downloaded files on your desktop 1. Delete the two downloaded zip files 1. Create a folder "'''brainstorm_db'''" on your '''Desktop ''' 1. Final check: you should have 3 folders on your desktop: * '''brainstorm3''': Program folder, with the source code and the compiled executable * '''brainstorm_db''': Brainstorm database (empty) * '''ds000117-practical''': Example dataset used during the training session 1. Start Brainstorm (see following section) 1. Select the menu '''Help > Workshop preparation'''. 1. Follow the instructions: when you see the 3D figure, make sure you see both a transparent head surface and a brain surface. Try to rotate the view with your mouse, and zoom in and out with the mouse wheel. <<BR>><<BR>> {{attachment:test_bst.gif}} 1. If you want to call FieldTrip functions from Brainstorm (multitaper, cluster-based statistics and dipole fitting), '''download and install FieldTrip''' on your computer:<<BR>>http://www.fieldtriptoolbox.org/download/ |
| Line 45: | Line 83: |
| <<HTML(<TR><TD>)>>11:00-12:30<<HTML(</TD><TD>)>>'''Practice''' | == Running Brainstorm for the first time == ==== With Matlab ==== 1. Matlab versions >= 2014b are faster and produce nicer graphics: consider upgrading if possible 1. Start Matlab 1. Do '''NOT '''add brainstorm3 folder to your Matlab path: this will be done automatically 1. Go to the brainstorm3 folder 1. Type "brainstorm" in the command window 1. When asked for the Brainstorm database folder, pick the "brainstorm_db" you have just created |
| Line 47: | Line 92: |
| Registration EEG/MRI | ==== Without Matlab ==== 1. The instructions below are valid for the following operating systems: * '''Windows''': Any version ([[https://www.mathworks.com/content/dam/mathworks/mathworks-dot-com/support/sysreq/files/SystemRequirements-Release2015b_Windows.pdf|details]]) * '''Linux''': Ubuntu 14.04+, RedHat 6.x+, Debian 7.x, SUSE 11.3+ ([[https://www.mathworks.com/content/dam/mathworks/mathworks-dot-com/support/sysreq/files/SystemRequirements-Release2015b_Linux.pdf|details]]) * '''MacOS''': 10.9.5 (Mavericks), 10.10 (Yosemite) ([[https://www.mathworks.com/content/dam/mathworks/mathworks-dot-com/support/sysreq/files/SystemRequirements-Release2015b_Macintosh.pdf|details]]) * If your system is not listed, try another version: [[InstallationR2012b|2012b]], [[InstallationR2013b|2013b]], [[InstallationR2014b|2014b]], [[InstallationR2016a|2016a]], [[InstallationR2016b|2016b]], [[InstallationR2017a|2017a]] 1. Download the '''MCR R2015b (9.0)''' for your operating system: [[http://www.mathworks.com/products/compiler/mcr/|Mathworks website]] 1. Install the MCR: * __Windows__: Double-click on the .exe and follow the instructions * __MacOS__: Click on the zip file to unzip it, then click on "InstallForMacOSX" * __Linux__: From a terminal, unzip .zip, then run ./install 1. Run the program in brainstorm3/bin/R2015b/ * __Windows__: Double-click on ''brainstorm3.bat'' * __MacOS__: Double-click on ''brainstorm3.command ''and wait for instructions * __Linux__: From a terminal, run:<<BR>>''cd brainstorm3/bin/R2015b/''<<BR>>''./brainstorm3.command '' 1. Troubleshooting for MacOS or Linux: * On recent versions of MacOS, you may get an error message "Application can't be opened because it is from an unidentified developer". This message would appear for all the programs that were not downloaded from the Apple app-store. To go around this verification: right-click on the application > Open, then click on the Open button. * From a terminal, make sure that the file "''brainstorm3.command''" is executable:<<BR>>''chmod a+x brainstorm3.command'' * If nothing happens, run: <<BR>>''./brainstorm3.command MCR_DIR''<<BR>>MCR_DIR is the MCR 9.0 folder (ex: /Applications/MATLAB/MCR/v90) * Try with another binary release: [[InstallationR2012b|2012b]], [[InstallationR2013b|2013b]], [[InstallationR2014b|2014b]], [[InstallationR2016a|2016a]], [[InstallationR2016b|2016b]], [[InstallationR2017a|2017a]]... 1. Troubleshooting for Windows: * Your current user may not have the necessary privileges. If you are an administrator for your computer, you can do the following: right-click on ''brainstorm3.bat'' > Run as administrator. * If you are not the administrator of your computer and Matlab or the MCR are not installed in the standard paths, Brainstorm may have trouble finding them. To specify manually the path of the MCR or Matlab folder, right-click on ''brainstorm3.bat'' > Edit. Fill the second line of the script (Example: @SET MATLABROOT="C:\Program Files\MATLAB\R2015b"), save the file, and try to execute it again. 1. On Linux or MacOS, you may be asked to select the folder where you installed the MCR. 1. When asked for the Brainstorm database folder, pick the "brainstorm_db" you have just created. |
| Line 49: | Line 118: |
| Review of continuous EEG recordings | == Bring your own data == The last session will be mostly dedicated to adressing participants' requests. We encourage you to prepare some EEG/MEG/ECoG/sEEG data you would be interested in processing with Brainstorm. If you are coming with colleagues, try to organize small groups with similar interests (1-4 people). |
| Line 51: | Line 121: |
| Frequency filters | The list of file formats that can be read by Brainstorm is available on the [[http://neuroimage.usc.edu/brainstorm/Introduction#Supported_file_formats|Introduction page]]. Please contact us in advance if you are not sure your dataset can be imported in Brainstorm. |
| Line 53: | Line 123: |
| Montages and bad channels <<HTML(</TD></TR>)>> <<HTML(<TR><TD>)>>12:30-13:30<<HTML(</TD><TD>)>>''Lunch''<<HTML(</TD></TR>)>> <<HTML(<TR><TD>)>>13:30-15:00<<HTML(</TD><TD>)>>'''Practice''' Artifact detection Artifact correction with SSP Averaging<<HTML(</TD></TR>)>> <<HTML(<TR><TD>)>>15:00-15:30<<HTML(</TD><TD>)>>''Coffee break''<<HTML(</TD></TR>)>> <<HTML(<TR><TD>)>>15:30-16:00<<HTML(</TD><TD>)>>'''Lecture: Source modeling (Baillet)'''<<HTML(</TD></TR>)>> <<HTML(<TR><TD>)>>16:00-17:30<<HTML(</TD><TD>)>>'''Practice''' Head modeling, cortical source reconstruction Regions of interest (ROIs) <<HTML(</TD></TR>)>> <<HTML(</TABLE>)>> == Program: Day 2 == <<HTML(<TABLE>)>> <<HTML(<TR><TD>)>>9:00-9:30<<HTML(</TD><TD>)>>'''Lecture: Time-frequency (Baillet)'''<<HTML(</TD></TR>)>> <<HTML(<TR><TD>)>>9:30-10:15<<HTML(</TD><TD>)>>'''Practice''' Time-frequency decompositions<<HTML(</TD></TR>)>> <<HTML(<TR><TD>)>>10:15-10:45<<HTML(</TD><TD>)>>'''Lecture: Connectivity & resting-state (Baillet)'''<<HTML(</TD></TR>)>> <<HTML(<TR><TD>)>>10:45-11:00<<HTML(</TD><TD>)>>''Coffee break''<<HTML(</TD></TR>)>> <<HTML(<TR><TD>)>>11:00-12:30<<HTML(</TD><TD>)>>'''Practice''' Connectivity tools Statistics Scripting<<HTML(</TD></TR>)>> <<HTML(<TR><TD>)>>12:30-13:30<<HTML(</TD><TD>)>>''Lunch''<<HTML(</TD></TR>)>> <<HTML(<TR><TD>)>>13:30-17:30<<HTML(</TD><TD>)>>'''Analyze your own data ''' Participants work in small groups (1-4 people) to analyze their own recordings<<HTML(</TD></TR>)>> <<HTML(</TABLE>)>> == Program: Day 3 == <<HTML(<TABLE>)>> <<HTML(<TR><TD>)>>9:00-14:00<<HTML(</TD><TD>)>>'''Analyze your own data ''' (continued) <<HTML(</TD></TR>)>> <<HTML(<TR><TD>)>>14:00-16:00<<HTML(</TD><TD>)>>'''What did you find?''' Participants present analysis results and impressions with the rest of the group<<HTML(</TD></TR>)>> <<HTML(</TABLE>)>> == Registration == To register to this event: [[https://www.labpsicologiaucm.com/news/workshop-brainstorm|Follow these instructions]] . <<EmbedContent(https://neuroimage.usc.edu/bst/workshop.js)>> . <<HTML(<!-- div id="bst_table" style=""><FORM METHOD="post" ACTION="https://neuroimage.usc.edu/bst/workshop_reg.php" name="form" onSubmit="return validateForm(this);"><TABLE><TR><TD align=right>First name * </TD> <TD><INPUT type="text" name="firstname" size=40></TD></TR> <TR><TD align=right>Last name * </TD> <TD><INPUT type="text" name="lastname" size=40></TD></TR> <TR><TD align=right>Title / Occupation * </TD> <TD><INPUT type="text" name="title" size=40></TD></TR> <TR><TD align=right>Institution * </TD> <TD><INPUT type="text" name="institution" size=40></TD></TR> <TR><TD align=right>City * </TD> <TD><INPUT type="text" name="city" size=40></TD></TR> <TR><TD align=right>Email address * </TD> <TD><INPUT type="text" name="email" size=40></TD></TR> <TR><TD align=right>Phone </TD> <TD><INPUT type="text" name="phone" size=40></TD></TR></TABLE> <BR> <BR> <INPUT type="submit" value="Register now!" style="width:200px"> <!-- A HREF="WorkshopCambridge2017Reg">View list of registered participants </A --> <BR><BR><BR><INPUT type="hidden" name="eventname" value="Cambridge2017"> </FORM> </div -->)>> |
If you are interested in using the subject's MRI for more realistic source estimation, you would have to process it in advance. You could try using the following programs: [[Tutorials/LabelFreeSurfer|FreeSurfer]], [[Tutorials/SegBrainVisa|BrainVISA]], [[Tutorials/SegBrainSuite|BrainSuite]], [[https://neuroimage.usc.edu/brainstorm/Tutorials/SegCAT12|CAT12]] |

Paris, France: December 3-5, 2019
Practical workshop on EEG/MEG analysis at ICM, in parallel with FieldTrip and MNE-Python. Program available on the workshop website: http://practicalmeeg2019.org/
The hands-on part of this workshop is based on the Frontiers Research Topic From raw MEG/EEG to publication: a collection of articles processing the same multimodal dataset using different software environments. We will process together one subject of this dataset, following the processing pipeline described in the article MEG/EEG Group Analysis With Brainstorm and the corresponding online tutorial.
General information
| Where | ICM |
| When | Tuesday Dec 3rd - Thursday Dec 5, 2019: 8:30-17:30 |
| Instructors | Francois Tadel (Grenoble Institute of Neuroscience) Anne-Sophie Dubarry (Aix-Marseille University) Aurélien Weiss (ENS) |
| Audience | Users interested in analyzing sEEG/EEG/ECoG/MEG recordings using Brainstorm. Teaching in English. |
| Documents | Introduction slides | Hands-on walkthrough | Processing script | Anatomy processing |
Requirements
The participants are required to bring a laptop (an external mouse will add to your comfort). In order to make the session as efficient as possible, we ask all the attendees to download, install and test the software and sample dataset on their laptops prior to the workshop.
Please read carefully the installation instructions:
?How to prepare your laptop for the training
Program
1. Tuesday am: From raw to ERP
- Introduction to Brainstorm (lecture)
- Review the MEG+EEG recordings
- Read and organize events
- Frequency filters
- Artifacts detection and cleaning
2. Tuesday pm: Sensor level analysis
- Epoching and averaging
- Time-frequency analysis
3. Wednesday am: Creating head and source models
- Import anatomy
- Registration MRI-sensors
- Forward model
- Simulations
4. Wednesday pm: Single and distributed sources
- MEG noise covariance: From empty room recordings
- EEG noise covariance: From pre-stimulus baselines
- Distributed sources / minimum norm estimation
- Regions of interest
- LCMV Beamformer
- Dipole scanning / Volume source models
5. Thursday am: Group-level analysis
- Project source maps on MNI template
- Statistical testing
Export data to other programs (FieldTrip, SPM, ...)
- Scripting
Example for this workshop: https://github.com/brainstorm-tools/brainstorm3/blob/master/toolbox/script/tutorial_practicalmeeg.m
6. Thursday pm: User requests
- Bring your own data or prepare your questions
Installation instructions
Before coming to the workshop, you need to download the software and the tutorial dataset (1.6 Gb). To streamline troubleshooting during the session, please save all the downloaded files on your Desktop.
From the Brainstorm Download page, log in or create a Brainstorm account
Download brainstorm_YYMMDD.zip (75 Mb)
If you already have Brainstorm: update it. We need a version released after November 18th
Download the tutorial dataset (1.5 Gb):
https://owncloud.icm-institute.org/index.php/s/cNu5jmiOhe7Yuoz/download- Unzip the two downloaded files on your desktop
- Delete the two downloaded zip files
Create a folder "brainstorm_db" on your Desktop
- Final check: you should have 3 folders on your desktop:
brainstorm3: Program folder, with the source code and the compiled executable
brainstorm_db: Brainstorm database (empty)
ds000117-practical: Example dataset used during the training session
- Start Brainstorm (see following section)
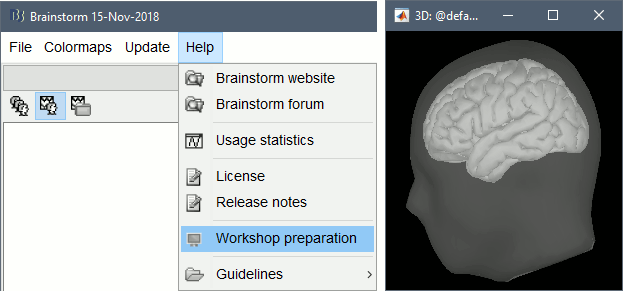
Select the menu Help > Workshop preparation.
Follow the instructions: when you see the 3D figure, make sure you see both a transparent head surface and a brain surface. Try to rotate the view with your mouse, and zoom in and out with the mouse wheel.
If you want to call FieldTrip functions from Brainstorm (multitaper, cluster-based statistics and dipole fitting), download and install FieldTrip on your computer:
http://www.fieldtriptoolbox.org/download/
Running Brainstorm for the first time
With Matlab
Matlab versions >= 2014b are faster and produce nicer graphics: consider upgrading if possible
- Start Matlab
Do NOT add brainstorm3 folder to your Matlab path: this will be done automatically
- Go to the brainstorm3 folder
- Type "brainstorm" in the command window
- When asked for the Brainstorm database folder, pick the "brainstorm_db" you have just created
Without Matlab
- The instructions below are valid for the following operating systems:
Download the MCR R2015b (9.0) for your operating system: Mathworks website
- Install the MCR:
Windows: Double-click on the .exe and follow the instructions
MacOS: Click on the zip file to unzip it, then click on "InstallForMacOSX"
Linux: From a terminal, unzip .zip, then run ./install
- Run the program in brainstorm3/bin/R2015b/
Windows: Double-click on brainstorm3.bat
MacOS: Double-click on brainstorm3.command and wait for instructions
Linux: From a terminal, run:
cd brainstorm3/bin/R2015b/
./brainstorm3.command
- Troubleshooting for MacOS or Linux:
On recent versions of MacOS, you may get an error message "Application can't be opened because it is from an unidentified developer". This message would appear for all the programs that were not downloaded from the Apple app-store. To go around this verification: right-click on the application > Open, then click on the Open button.
From a terminal, make sure that the file "brainstorm3.command" is executable:
chmod a+x brainstorm3.commandIf nothing happens, run:
./brainstorm3.command MCR_DIR
MCR_DIR is the MCR 9.0 folder (ex: /Applications/MATLAB/MCR/v90)Try with another binary release: 2012b, 2013b, 2014b, 2016a, 2016b, 2017a...
- Troubleshooting for Windows:
Your current user may not have the necessary privileges. If you are an administrator for your computer, you can do the following: right-click on brainstorm3.bat > Run as administrator.
If you are not the administrator of your computer and Matlab or the MCR are not installed in the standard paths, Brainstorm may have trouble finding them. To specify manually the path of the MCR or Matlab folder, right-click on brainstorm3.bat > Edit. Fill the second line of the script (Example: @SET MATLABROOT="C:\Program Files\MATLAB\R2015b"), save the file, and try to execute it again.
- On Linux or MacOS, you may be asked to select the folder where you installed the MCR.
- When asked for the Brainstorm database folder, pick the "brainstorm_db" you have just created.
Bring your own data
The last session will be mostly dedicated to adressing participants' requests. We encourage you to prepare some EEG/MEG/ECoG/sEEG data you would be interested in processing with Brainstorm. If you are coming with colleagues, try to organize small groups with similar interests (1-4 people).
The list of file formats that can be read by Brainstorm is available on the Introduction page. Please contact us in advance if you are not sure your dataset can be imported in Brainstorm.
If you are interested in using the subject's MRI for more realistic source estimation, you would have to process it in advance. You could try using the following programs: FreeSurfer, BrainVISA, BrainSuite, CAT12
