Hello everyone,
I want to use brainstorm to process ADHD EEG data. The participants were young children (8-12 years), but I do not have individual head model. I wonder whether there are common head model for kids?
Thank you so much!
Best,
Yu
Hello everyone,
I want to use brainstorm to process ADHD EEG data. The participants were young children (8-12 years), but I do not have individual head model. I wonder whether there are common head model for kids?
Thank you so much!
Best,
Yu
Hi Yu,
In Brainstorm, there are only templates for babies readily available (7 weeks, 1 year), which are not going to match the age range you need:
https://neuroimage.usc.edu/brainstorm/Tutorials/DefaultAnatomy#BrainVISA_templates
This is a question we get regularly on this forum, some of these discussions might help you, or maybe try to contact the users who posted the questions directly:
The ideal would be to have a digitized shape of the head shape of all the children, you could use this head shape to warp the adult template ICBM152:
https://neuroimage.usc.edu/brainstorm/Tutorials/TutWarping
Another solution would be to find templates for the ages you are interested in (the are probably some available publicly now), and process them with FreeSurfer/BrainVISA/BrainSuite, to then be able to import them into Brainstorm as the subject's anatomy.
If you go for the latter solution, please let us know where you found the templates, this would be very useful for our users. If you have time for this, please also share with us the segmentation you obtained for these templates, we could include them as new templates available in Brainstorm. This contribution would be very much appreciated by many of our our users working with children EEG/MEG recordings.
Cheers,
Francois
Much appreciated, Francois! I will keep you posted if there are any further updates.
I have another question. Will it be possible to use brainstorm to perform time-frequency transform in all trials and all subjects first, and then grand average the data in frequency domain across subjects? And finally do source localization as well as functional connectivity analyses. Could you please kindly recommend any manuals to conduct the processing?
Thank you very much!
Best,
Yu
Start by following all the introduction tutorials (tutorial #27 gives our recommended processing pipelines for group analysis):
https://neuroimage.usc.edu/brainstorm/Tutorials
Another example for group analysis is available in the advanced tutorials:
https://neuroimage.usc.edu/brainstorm/Tutorials/VisualSingle
https://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup
Many thanks, Francois!
The data I processed is only phase-locked, not ERPs (both time-locked and phase-locked), so I want to:
However, I found that I cannot directly do source localization in the grand-averaged TF data ( frequency damain). So I tried to 1)compute source in each participant, and 2) then drag all participants data and follow the scout guide here:https://neuroimage.usc.edu/brainstorm/Tutorials/TimeFrequency. I used D-K scouts and frequency bands ( alpha, beta,etc). But 2) took really long. I wonder whether the process is correct. If it is not, could you please kindly help me?
Thank you very much!
So I tried to 1)compute source in each participant, and 2) then drag all participants data and follow the scout guide here:https://neuroimage.usc.edu/brainstorm/Tutorials/TimeFrequency.
Indeed, what you describe is similar to what is illustrated in the "Scout" section of this tutorial. Make sure that when you run this, you really select all the individual trials and not the average. Repeat for each subject and then average across subjects.
The recommend order for the different steps is described here:
https://neuroimage.usc.edu/brainstorm/Tutorials/Workflows#Time-frequency_maps
I used D-K scouts
Be careful, this is an anatomical atlas, with very inhomogeneous ROI sizes, not very well suited for functional analysis. If you have strong hypotheses in your analysis, it would be better to design your own ROIs.
But 2) took really long
Working in source space, with 15000 signals instead of a few tens of sensors, is painful, indeed...
If this is not manageable, you could try averaging the scouts signals before computing the TF decomposition (option "Scout function: Before" in the TF process options). Make sure it does not degrade too much your results first.
Thank you so much, Francois!
I did source-level time-frequency transformation in each subject as you advice, and got 'Avg,Power,FreqBands' under each subject. Then I put all the 'Avg,Power,FreqBands' files in Process1, and tried to average (Group files: everything). However, 1 warning occurred as follows. Could you please kind tell me how to correctly do the average? Many thanks!
I have another question, I supposed to see motor/sensorimotor ERD, but the activation region (attached) seemed to be a bit down than expected. I used Neuroscan 47 channels cap, but in source localization process, there was no suitable file (add EEG positions). So I did not load any file in add EEG position. Did this affect the ERD results? Could you please tell me how to correct this error? Thank you so much!
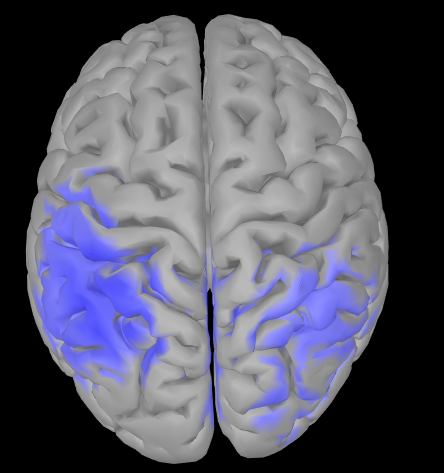
You need to verify and adjust (manually) the channel locations on scalp: if they are off with respect to the actual positions on your participant(s), this would explain the offset in the source maps.
Right click over the channel file and select "check registration" see
https://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Manual_registration
I did source-level time-frequency transformation in each subject as you advice, and got 'Avg,Power,FreqBands' under each subject. Then I put all the 'Avg,Power,FreqBands' files in Process1, and tried to average (Group files: everything). However, 1 warning occurred as follows. Could you please kind tell me how to correctly do the average?
If all your source maps were computed on the same template brain, you can safely ignore this warning as the order of the sources is the same in all your time-frequency files. As suggested by the warning, you can simply disable the option "Match signals between files using their names".
I used Neuroscan 47 channels cap, but in source localization process, there was no suitable file (add EEG positions).
I never saw references to this 47 channel cap.
If the data channels named with the 10-10 convention (Fp1, Cz, etc), you can maybe use any of the 10-10/10-20/10-05 templates (from the Generic menu, or the Neuroscan MagLink cap).
Otherwise, you'd need a proper template for this. Could you try finding standard 3D positions for your EEG cap, or ask them directly to the Neuroscan customer support? We could add them to Brainstorm.
So I did not load any file in add EEG position. Did this affect the ERD results?
If you don't set 3D positions for your electrodes, you cannot expect to obtain anything meaningful from the source reconstruction. If what you used is the flat 2D positions included in the Neuroscan files, you can discard all your source analysis.
Start by making sure your electrodes are correctly placed on the head surface:
https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy#Register_electrodes_with_MRI
Thank you so much, Francois!
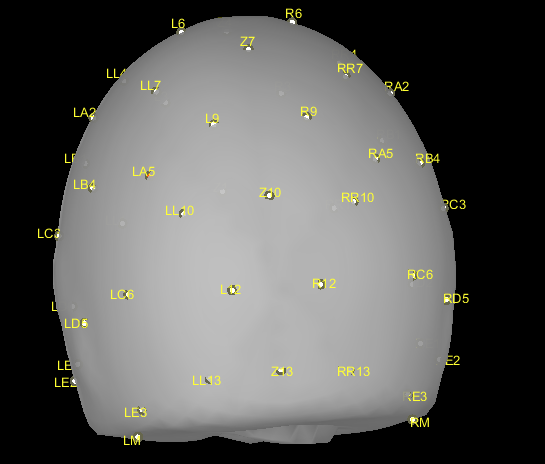
I loaded the 47-channel location file (attached) in the preprocessing at the beginning. But all the electrodes were a bit down (attached Capture2), e.g., some electrodes were on the bottom, some were even outside of the head. Could you please kindly help me to move up all electrodes a bit?
Much appreciated!
Best,
Yu
newelecfile.locs (1.4 KB)
 EEProbe continuous data epochs resampledP002_4_downSampling.set (82.2 KB)
EEProbe continuous data epochs resampledP002_4_downSampling.set (82.2 KB)
The file newelecfile.locs you posted do not contain any 3D electrodes positions. What you have in there are flat 2D positions that can be used only for a flat topography display in EEGLAB. You cannot expect to do any source reconstruction directly with these positions.
The positions you get when importing your EEGLAB file into Brainstorm are spherical projections of these 2D positions (you have a warning telling you this).
If you want to use these default spherical positions, you need to edit them manually:
With spherical positions, never try anything like warping the standard anatomy on the sphere or adjusting automatically the electrodes on the head surface. There is too much information you don't have and we can't reconstruct for you, you have to place your electrodes manually on the head.
Thank you so much for your reply, Francois!
I did have an warning when importing my EEGLAB file (attached warning1), and I clicked yes.
what I did was as follows. I am worried that I did not actually conduct source reconstruction. Could you please tell whether or not I did the source localization? Thanks a lot!
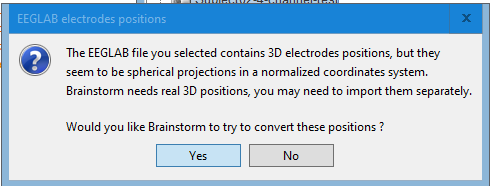
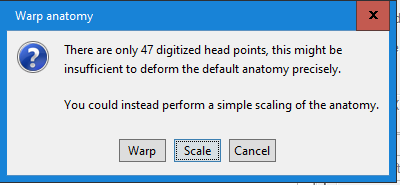
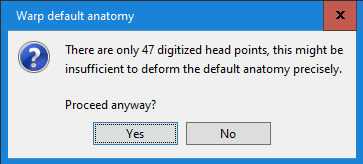
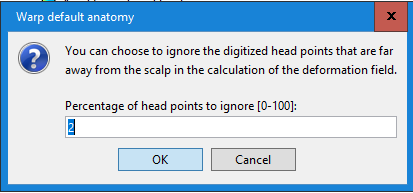
No, as I was mentioning in my previous message, you should use this warping option only when you have real digitized head points. Here what you did was to project transform the MNI brain into a perfect sphere...
You should go the other way around: do not alter the MNI template, and project the electrodes on the head surface.
Got it. Thank you very much!
One more question, I checked the location in EEGLAB, and there were X,Y,Z coordinates for each electrode. I can also plot 3-D(xyz). Could you please tell what the 3D electrodes positions file in brainstorm looks like? Thanks!
These are spherical coordinates (=projected to a sphere), not realistic digitized ones.
Thanks, Francois!
An engineer in ANT customer service gave me a new channel location file (please see attached). Could you please help me check it a realistic digitized one or not ? Thank you!
Standard_47chans.elc (1.3 KB)
This looks better.
Is this a standard cap sold by Neuroscan?
Is so, I could prepare a new Brainstorm MNI template for it.
What is the full commercial name of this EEG cap?
Can you point at some documentation mentioning it?
Do you have some pictures that show people with the cap on their so?
I would need to know more or less how it is supposed to fit on the head (how far the frontal electrodes are supposed to be from the eyebrows, how far do they go down the neck, how do they fit around the ears)
Thanks
Btw, I fixed a bug that existed when reading the electrode names from this .elc files: https://github.com/brainstorm-tools/brainstorm3/commit/d43d81462c2d29a83bc8b7f1a6f61e0c0ee4f008
Update Brainstorm to get this fix.