|
Size: 5131
Comment:
|
Size: 18694
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Tutorial 11: Averaging = | = Tutorial 16: Average response = |
| Line 3: | Line 3: |
All the epochs we have imported in the previous tutorial are represented by matrices that have the same size (same number of channels, same number of time points), therefore they can be averaged together by experimental condition. The result is called indifferently "'''evoked response'''", "'''average response'''", "'''event-related field'''" in MEG (ERF) or "'''event-related potential'''" in EEG (ERP). It shows the components of the brain signals that are strictly time-locked to the presentation of a stimulus. . {{attachment:average_slide.gif||height="185",width="390"}} |
|
| Line 6: | Line 10: |
| = From auditory = === Average responses === * As said previously, it is usually not recommended to average recordings in sensor space across multiple acquisition runs because the subject might have moved between the sessions. Different head positions were recorded for each run, we will reconstruct the sources separately for each each run to take into account those movements. * However, in the case of event-related studies it makes sense to start our data exploration with an average across runs, just to evaluate the quality of the evoked responses. We have seen that the subject almost didn't move between the two runs, so the error would be minimal. We will compute now an approximate sensor average between runs, and we will run a more formal average in source space later. * We have 80 good "deviant" trials that we want to average together. * Select the trial groups "deviant" from both runs in Process1, run process "'''Average > Average files'''" Select the option "'''By trial group (subject average)'''" |
== Averaging == We will now compute the average responses for both the "standard" and "deviant" conditions. Two constraints have to be taken into consideration at this stage. |
| Line 14: | Line 13: |
| {{http://neuroimage.usc.edu/brainstorm/Tutorials/Auditory?action=AttachFile&do=get&target=process_average_data1.gif|process_average_data1.gif|height="456",width="470",class="attachment"}} * To compare properly this "deviant" average with the other condition, we need to use the same number of trials in the "standard" condition. We are going to pick 40 "standard" trials from Run01 and 40 from Run02. To make it easy, let's take the 40 first good trials. * Select the '''41 '''first "standard" trials of Run01 + the '''41 '''first "standard" trials of Run02 in Process1. This will sum to '''80 '''selected files, because the Process1 tab ignores the bad trials (trial #37 is bad in Run01, trial #36 is bad in Run02) * Run again process "'''Average > Average files'''" > "'''By trial group (subject average)'''" |
'''Averaging runs separately''': With MEG, it is not recommended to average sensor data across acquisition runs with different head positions (ie. different "channel files"). If the head of the subject moved between two blocks of recordings, a sensor does not record the same part of the brain before and after, therefore the runs cannot be compared directly. With EEG, you can generally skip this recommendation. |
| Line 20: | Line 15: |
| {{http://neuroimage.usc.edu/brainstorm/Tutorials/Auditory?action=AttachFile&do=get&target=process_average_data2.gif|process_average_data2.gif|height="444",width="471",class="attachment"}} * The average for the two conditions "standard" and "deviant" are saved in the folder ''(intra-subject)''. The channel file added to this folder is an average of the channel files from Run01 and Run02. |
'''Number of trials''': When computing subject-level averages for experimental conditions with different number of trials, you have two options. You can either use the same number of trials for all the conditions and subjects (to make them "more comparable") or to use all the good trials even there is a serious imablance in the experimental design (more samples always lead to better estimators for the mean and variance). Here we will go for the second option, using '''all the trials'''. However, in some specific cases it might not be the best choice, see the advanced section [[http://neuroimage.usc.edu/brainstorm/Tutorials/Averaging#Number_of_trials|Number of trials]] for additional details. |
| Line 23: | Line 17: |
| {{http://neuroimage.usc.edu/brainstorm/Tutorials/Auditory?action=AttachFile&do=get&target=average_sensor_files.gif|average_sensor_files.gif|height="205",width="220",class="attachment"}} | The instructions below show how to easily select an equal number of trials from the database: |
| Line 25: | Line 19: |
| = From continuous tutorials: = == Averaging == Drag and drop the two lists of trials (or the two condition folders) in the Process1 list, and run the process "'''Average > Average files'''", with the option "'''Average by condition (subject average)'''". This will group the trials by conditions, and calculate one average per condition and subject. The option "By condition (grand average)" would average each condition for all the subjects together, but as there is only one subject in this case, the result would be same. The other options would generate one average file per subject ("by subject"), one average file per group of trials and per subject ("by trial group"), or only one average no matter what the input is ("everything"). |
* Drag and drop all the "'''standard'''" and "'''deviant'''" trials for '''both runs''' in Process1. * In the Process1 list, you will notice that the number of imported trials (comment in the database explorer, eg. "40 files") does not match the number of files selected for processing (between brackets, eg. "[39]"). This difference is due to the '''bad trials''' that we have in these folders. The trials tagged with a red dot in the database explorer are ignored by all the processes. The total number of selected files is 468 instead of 480, it means that we have a total of 12 bad trials. * Select the process "'''Average > Average files'''".<<BR>>Select the options: '''By trial group (folder)''', Arithmetic average, Keep all the event markers.<<BR>><<BR>> {{attachment:average_folder.gif||height="547",width="525"}} |
| Line 29: | Line 23: |
| The function to apply is the regular arithmetic average. The option "'''Keep all the events from the individual epochs'''" would group all the event markers present in all the epochs and save them in the new averaged file. It can be useful to check the relative position of the artifacts or the subject responses, or quickly detect some unwanted configuration such as a subject who would constantly blink right after a visual stimulus. We don't really need this here, leave this option '''unselected'''. | * You get two new files for each average. The number of between parenthesis indicates the number of good trials that were used to compute each file. <<BR>><<BR>> {{attachment:average_files.gif||height="229",width="250"}} |
| Line 31: | Line 25: |
| {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=avgOptions.gif|avgOptions.gif|class="attachment"}} | == Process options: Average == Description of all the options of the process: Average > Average files. |
| Line 33: | Line 28: |
| It creates two files: "'''Avg: left'''" and "'''Avg: right'''". | * '''Everything''': Averages all the files selected in Process1 together, creates only one file in output. * '''By subject''': Groups the files by subject (ignoring the folders), creates one file per subject. * '''By folder (subject average)''': Groups by subject and by folder, ignoring the trial groups. <<BR>>In the current configuration, it would produce two files, one for each acquisition run. * '''By folder (grand average)''': Groups by folder, across the subjects. All the files located in folders with the same name would be averaged together, no matter in what subject they are. * '''By trial group (folder average)''': Groups by set of trials with the same name, separately for each folder and each subject. * '''By trial group (subject average)''': Groups by set of trials with the same name, for each subject. The separation in folders is ignored. In the current configuration, it would produce two files, one for "deviant" and one for "standard". * '''By trial group (grand average)''': Groups by set of trials with the same name, ignoring the classification by folder or subject. * '''Function''': Documented directly in the option panel. * '''Weighted average''': When averaging single trials, the number of files is saved in the field nAvg of the average file. When re-averaging the averages across acquisition sessions or subjects, this field nAvg can be used to weight each file with the number of trials from which it was computed: <<BR>>mean(x) = sum(nAvg(i) * x(i)) / sum(nAvg(i))<<BR>>In most cases, this option should be selected when averaging within a subject and disabled when averaging across subjects. It has no impact in the current example (no averages, nAvg=1). * '''Keep all the event markers''': If this option is selected, all the event markers that were available in all the individual trials are reported to the average file. It can be useful to check the relative position of the artifacts or the subject responses, or quickly detect some unwanted configuration such as a subject who would constantly blink immediately after a visual stimulus. |
| Line 35: | Line 39: |
| {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=avgDb.gif|avgDb.gif|class="attachment"}} | == Visual exploration == The average response contains interesting information about the brain operations that occur shortly after the presentation of the stimulus. We can explore two dimensions: the '''location''' of the various brain regions involved in the sensory processing and the precise '''timing''' of their activation. Because these two types of information are of equal interest, we typically explore the recordings with two figures at the same time, one that shows all the signals in time, one that shows the spatial distribution at one instant. |
| Line 37: | Line 42: |
| Double-click on the "'''Avg: Left'''" file to display the MEG recordings for this file (or right-click > MEG > display time series). It shows a very typical an clean evoked response, with a very high signal-to-noise ratio. | * Open the MEG recordings for the '''deviant average''' in '''Run#01''': double-click on the file. * In the Record tab: Select the "butterfly" view more (first button in the toolbar). * In the Filter tab: Add a '''low-pass filter''' at '''100Hz'''. * In the Record tab: Delete the "cardiac" event type, we are not interested by their distribution.<<BR>><<BR>> {{attachment:deviant_ts.gif||height="176",width="661"}} * This figure shows a typical clean evoked response, with a high signal-to-noise ratio. This represents the brain response to a simple auditory stimulation, the large peak around '''90ms''' probably corresponds to the main response in the primary auditory cortex. * The green line represents the global field power ('''GFP'''), ie the sum of the square of all the sensors at each time point. This measure is sometimes used to identify transient or stable states in ERP/ERF. * This is the response to the deviant beeps (clearly higher in pitch), for which the subject is supposed to press a button to indicate that he/she detected the target. These responses are represented with the "button" events, distributed between 350ms and the end of the file (many responses happened after 500ms). Because of the variability in the response times, we can already anticipate that we won't be able to study correctly the motor response from this average. For studying the activity in the motor area, we need to epoch the recordings again around the "button" events. |
| Line 39: | Line 50: |
| {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=avgTs.gif|avgTs.gif|class="attachment"}} | Add a spatial view: |
| Line 41: | Line 52: |
| Just for the records, this is how it would look like if we had selected the option "Keep all the events from the individual epochs". The summary of the markers of all the epochs is: 37 heartbeats and 2 blinks. | * Open a 2D topography for the same file (right-click on the figure > View topography, or Ctrl+T). * Review the average '''as a movie''' with the keyboard shortcuts (hold the left or right arrow key). * At '''90ms''', we can observe a typical topography for a bilateral auditory response. Both on the left sensors and the right sensors we observe field patterns which seem to indicate a dipolar-like activity in the temporal or central regions. <<BR>><<BR>> {{attachment:deviant_topo.gif||height="136",width="159"}} * Close everything with the button [X] in the top-right corner of the Brainstorm window. <<BR>>Accept to save the modifications (you deleted the "cardiac" events). * Open the "standard" average in the same way and delete the "cardiac" markers. |
| Line 43: | Line 58: |
| {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=avgTsEvents.gif|avgTsEvents.gif|class="attachment"}} | Repeat the same operations for '''Run#02''': * Open the MEG recordings for deviant and standard. * Delete the "cardiac" markers in both files. * Open a 2D topography and review the recordings. * Close everything. == Interpretation == Let's display the two conditions "'''standard'''" and "'''deviant'''" side-by-side, for '''Run#01'''. * Right-click on average > MEG > Display time series. * Right-click on average > MISC > Display time series (EEG electrodes Cz and Pz) * Right-click on average > MEG > 2D Sensor cap * In the Filter tab: add a '''low-pass filter''' at '''40Hz'''. * In the Record tab: you can set a common amplitude scale for all the figures with the button '''[=]'''. * Here are the results for the standard (top) and deviant (bottom) beeps: <<BR>><<BR>> {{attachment:average_summary.gif||width="650"}} The legend in blue shows names often used in the EEG ERP literature: * '''P50''': 50ms, bilateral auditory response in both conditions. * '''N100''': 95ms, bilateral auditory response in both conditions. * '''MMN''': 230ms, mismatch negativity in the deviant condition only (detection of an abnormality). * '''P200''': 170ms, in both conditions but much stronger in the standard condition. * '''P300''': 300ms, deviant condition only (decision making in preparation of the button press). * Some seem to have a direct correspondance in MEG (N100) some don't (P300). Additional quality check with the event markers: * The time series figure for the standard condition show two unwanted events between 100ms and 200ms post-stimulus, one "blink" and one "button" response. The trials that contain them should be marked as bad and the average recomputed, because the subject is not doing the task correctly. * We will not do it here, but keep in mind that this option "Keep all events" from the averaging process give a good summary view of the recordings and can help you identify some bad trials. <<TAG(Advanced)>> == Averaging bad channels == The bad channels can be defined independently for each trial and each channel is averaged independently, therefore we can have different numbers of data points averaged for different electrodes. If we have a channel A considered good for N<<HTML(<SUB>A</SUB>)>> trials, the corresponding channel in the average file is: sum(N<<HTML(<SUB>A</SUB>)>> trials) / N<<HTML(<SUB>A</SUB>)>>. In the averaged file, a channel is consider as good if it is good in at least one trial, and considered as bad if it is bad in all the trials. The entire file is then considered as if it were computed from N<<HTML(<SUB>max</SUB>)>> trials, N<<HTML(<SUB>max</SUB>)>> = max(N<<HTML(<SUB>i</SUB>)>>, i=1..N<<HTML(<SUB>trials</SUB>)>>. This may cause some unwanted effects across channels: the SNR might be higher for some channels than others. This way of proceeding was decided to allow the conservation of the maximum amount of data. If you want to avoid this to happen: mark the channels as bad in all the trials, or report all the bad channels to the average file. This can be done easily using the database explorer, see tutorial [[Tutorials/BadChannels|Bad channels]]. <<TAG(Advanced)>> == Averaging across runs == As said previously, it is usually not recommended to average recordings in sensor space across multiple acquisition runs because the subject might have moved between the sessions. Different head positions were recorded for each run, so we will reconstruct the sources separately for each each run to take into account these movements. However, in the case of event-related studies it makes sense to start our data exploration with an average across runs, just to evaluate the quality of the evoked responses. We have seen in tutorial #4 that the subject almost didn't move between the two runs, so the error would be minimal. . {{http://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile?action=AttachFile&do=get&target=channel_multiple.gif|channel_multiple.gif|height="218",width="438",class="attachment"}} Let's compute an approximate average across runs. We will run a formal average in source space later. * To run the same process again with different parameters: '''File > Reload last pipeline'''. Select: * '''By trial group (subject average)''': It will compute one average per experimental condition. * '''Arithmetic average + Standard error''': It will save, in the same file, the average and the standard error across all the trials, illustrated in the next section. * '''Keep all the event markers''': Do not select this option, we've already seen what it does. * The two files that are created are now saved in a new folder "'''Intra-subject'''". This is where all the results of processes involving multiple folders, within one subject, will be saved.<<BR>><<BR>> {{attachment:average_stderror.gif||height="184",width="219"}} <<TAG(Advanced)>> == Standard error == If you computed the '''standard deviation''' or the '''standard error''' together with an average, it will be automatically represented in the time series figures. * Double-click on one of the AvgStderr files to display the MEG sensors.<<BR>>The light-grey area around the sensors represent the maximum standard error around the maximum and minimum values across all the sensors. * Select two sensors and plot them separately (right-click > Channels > View selected, or "Enter").<<BR>>The green and red areas represent, at each time point, the standard error around the signal.<<BR>><<BR>> {{attachment:stderror.gif||height="176",width="379"}} * Right-click on the file > '''File > View file contents'''. <<BR>>The average is saved in the field '''F''', the standard error is saved in the field '''Std'''.<<BR>><<BR>> {{attachment:stderror_file.gif||height="230",width="396"}} <<TAG(Advanced)>> == Bad channels == <<TAG(Advanced)>> == Number of trials == You should always be careful when comparing averages computed from different numbers of trials. In most cases, you can safely include '''all the trials''' in your averages, even in the case of imbalanced designs. However, for very low numbers of trials, having the same number of trials becomes more critical. <<BR>>See the following references for more details: * Luck SJ (2010)<<BR>>[[http://erpinfo.org/educational-materials/comments-on-comparing-conditions-with-different-numbers-of-trials-1|Is it Legitimate to Compare Conditions with Different Numbers of Trials?]] * Thomas DG, Grice JW, Najm-Briscoe RG, Miller JW (2004)<<BR>>[[http://www.ncbi.nlm.nih.gov/pubmed/15525568|The influence of unequal numbers of trials on comparisons of average event-related potentials]] <<TAG(Advanced)>> == Selecting the trials == We will now compute the average responses for both the "standard" and "deviant" conditions. Two constraints have to be taken into consideration at this stage. '''Using the same number of trials''': As the goal in this analysis is to directly compare the two experimental conditions, we should use the same number of trials for computing the averages. We need to find the group of trials with the lowest number of good trials, and pick the same number in every group. Here we will be limited to 39 trials because of the deviant condition in Run#01. If we had multiple subjects and were planning to compute some group statistics, we should also use equal numbers of trials for all the subjects. The instructions below show how to easily select an equal number of trials from the database: * Drag and drop all the "'''standard'''" and "'''deviant'''" trials for '''both runs''' in Process1. * Select the process "'''Files > Select uniform number of trials'''".<<BR>>Select the options: '''By trial group (folder)''' and '''Uniformly distributed'''.<<BR>><<BR>> {{attachment:select_process.gif||height="394",width="532"}} * If you click on [Run], it doesn't do anything but highlighting the first selected file in the database explorer. This process just performs a file selection, it needs to be followed by another process that uses the selected files for computing something. However, you can see what was done in the process report. The reports are displayed only when an error or a warning was reported, but you can open them manually to check for additional messages. Menu '''File > Report viewer'''.<<BR>><<BR>> {{attachment:select_report.gif||height="226",width="633"}} * The comment in the report shows the 4 groups of trials that were identified based on the option we selected (group "by trial group and folder"), with the number of good trials per group: <<BR>>Run01/deviant (39), Run01/standard (193), Run02/deviant (40), Run01/standard (197).<<BR>>The process picked 39 trials in each group, uniformly distributed in the list of available trials. <<BR>>Example of trial indices selected for Run01/standard: [1, 6, 11, 16, 21, 26, 31, 36, ..., 188, 193] <<TAG(Advanced)>> == Process options: Select trials == Available options in the process: File > Select uniform number of trials. * '''By folder''': Groups by subject and by folder, ignoring the trial groups. <<BR>>Here, it would identify two groups, one for each acquisition run: Run01, Run02. * '''By trial group (folder)''': Groups by set of trials with the same name, separately for each folder and each subject. Here it would identify four groups: Run01/deviant, Run01/standard, Run02/deviant, Run01/standard. * '''By trial group (subject)''': Groups by set of trials with the same name, for each subject. The separation in folders is ignored. Here it would identify two groups: deviant, standard. How to select trials in a group that contains more than the requested number (Nf files, selecting only Ns): * '''Random selection''': Select a random subset of Ns trials. Trial indices: randperm(Nf,Ns) * '''First in the list''': Select the first Ns trials. Trial indices: 1:Ns * '''Last in the list''': Select the last Ns trials. Trial indices: Nf-Ns+1:Nf * '''Uniformly distributed''': Select Ns equally spaced trials. Trial indices: round(linspace(1, Nf, Ns))) <<HTML(<!-- END-PAGE -->)>> |
Tutorial 16: Average response
Authors: Francois Tadel, Elizabeth Bock, Sylvain Baillet
All the epochs we have imported in the previous tutorial are represented by matrices that have the same size (same number of channels, same number of time points), therefore they can be averaged together by experimental condition. The result is called indifferently "evoked response", "average response", "event-related field" in MEG (ERF) or "event-related potential" in EEG (ERP). It shows the components of the brain signals that are strictly time-locked to the presentation of a stimulus.
Contents
Averaging
We will now compute the average responses for both the "standard" and "deviant" conditions. Two constraints have to be taken into consideration at this stage.
Averaging runs separately: With MEG, it is not recommended to average sensor data across acquisition runs with different head positions (ie. different "channel files"). If the head of the subject moved between two blocks of recordings, a sensor does not record the same part of the brain before and after, therefore the runs cannot be compared directly. With EEG, you can generally skip this recommendation.
Number of trials: When computing subject-level averages for experimental conditions with different number of trials, you have two options. You can either use the same number of trials for all the conditions and subjects (to make them "more comparable") or to use all the good trials even there is a serious imablance in the experimental design (more samples always lead to better estimators for the mean and variance). Here we will go for the second option, using all the trials. However, in some specific cases it might not be the best choice, see the advanced section Number of trials for additional details.
The instructions below show how to easily select an equal number of trials from the database:
Drag and drop all the "standard" and "deviant" trials for both runs in Process1.
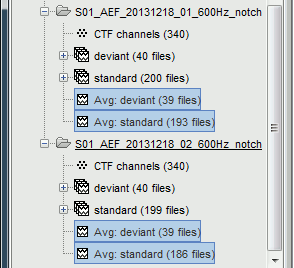
In the Process1 list, you will notice that the number of imported trials (comment in the database explorer, eg. "40 files") does not match the number of files selected for processing (between brackets, eg. "[39]"). This difference is due to the bad trials that we have in these folders. The trials tagged with a red dot in the database explorer are ignored by all the processes. The total number of selected files is 468 instead of 480, it means that we have a total of 12 bad trials.
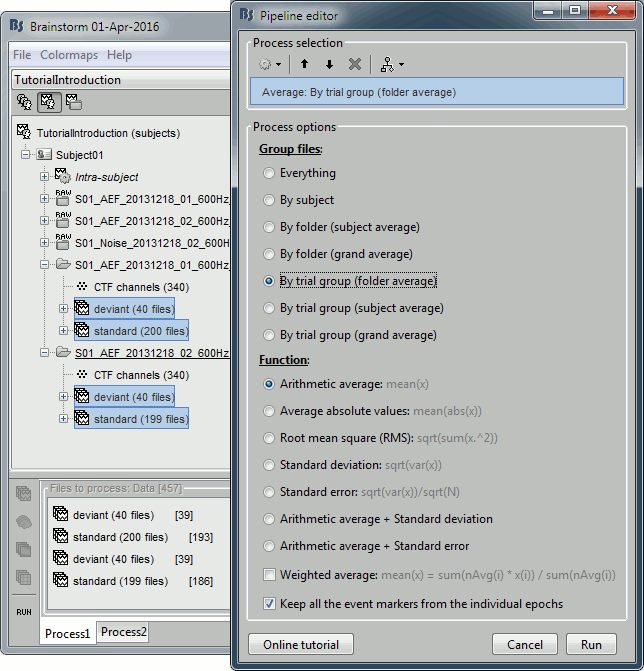
Select the process "Average > Average files".
Select the options: By trial group (folder), Arithmetic average, Keep all the event markers.
You get two new files for each average. The number of between parenthesis indicates the number of good trials that were used to compute each file.
Process options: Average
Description of all the options of the process: Average > Average files.
Everything: Averages all the files selected in Process1 together, creates only one file in output.
By subject: Groups the files by subject (ignoring the folders), creates one file per subject.
By folder (subject average): Groups by subject and by folder, ignoring the trial groups.
In the current configuration, it would produce two files, one for each acquisition run.By folder (grand average): Groups by folder, across the subjects. All the files located in folders with the same name would be averaged together, no matter in what subject they are.
By trial group (folder average): Groups by set of trials with the same name, separately for each folder and each subject.
By trial group (subject average): Groups by set of trials with the same name, for each subject. The separation in folders is ignored. In the current configuration, it would produce two files, one for "deviant" and one for "standard".
By trial group (grand average): Groups by set of trials with the same name, ignoring the classification by folder or subject.
Function: Documented directly in the option panel.
Weighted average: When averaging single trials, the number of files is saved in the field nAvg of the average file. When re-averaging the averages across acquisition sessions or subjects, this field nAvg can be used to weight each file with the number of trials from which it was computed:
mean(x) = sum(nAvg(i) * x(i)) / sum(nAvg(i))
In most cases, this option should be selected when averaging within a subject and disabled when averaging across subjects. It has no impact in the current example (no averages, nAvg=1).Keep all the event markers: If this option is selected, all the event markers that were available in all the individual trials are reported to the average file. It can be useful to check the relative position of the artifacts or the subject responses, or quickly detect some unwanted configuration such as a subject who would constantly blink immediately after a visual stimulus.
Visual exploration
The average response contains interesting information about the brain operations that occur shortly after the presentation of the stimulus. We can explore two dimensions: the location of the various brain regions involved in the sensory processing and the precise timing of their activation. Because these two types of information are of equal interest, we typically explore the recordings with two figures at the same time, one that shows all the signals in time, one that shows the spatial distribution at one instant.
Open the MEG recordings for the deviant average in Run#01: double-click on the file.
- In the Record tab: Select the "butterfly" view more (first button in the toolbar).
In the Filter tab: Add a low-pass filter at 100Hz.
In the Record tab: Delete the "cardiac" event type, we are not interested by their distribution.
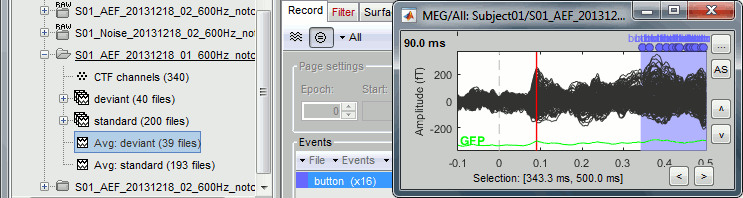
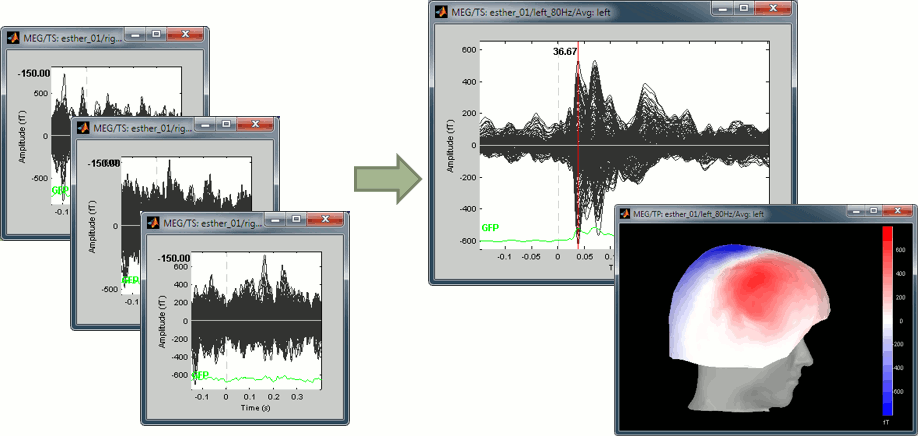
This figure shows a typical clean evoked response, with a high signal-to-noise ratio. This represents the brain response to a simple auditory stimulation, the large peak around 90ms probably corresponds to the main response in the primary auditory cortex.
The green line represents the global field power (GFP), ie the sum of the square of all the sensors at each time point. This measure is sometimes used to identify transient or stable states in ERP/ERF.
- This is the response to the deviant beeps (clearly higher in pitch), for which the subject is supposed to press a button to indicate that he/she detected the target. These responses are represented with the "button" events, distributed between 350ms and the end of the file (many responses happened after 500ms). Because of the variability in the response times, we can already anticipate that we won't be able to study correctly the motor response from this average. For studying the activity in the motor area, we need to epoch the recordings again around the "button" events.
Add a spatial view:
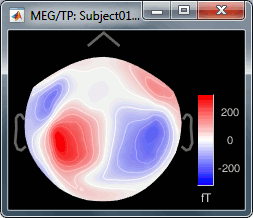
Open a 2D topography for the same file (right-click on the figure > View topography, or Ctrl+T).
Review the average as a movie with the keyboard shortcuts (hold the left or right arrow key).
At 90ms, we can observe a typical topography for a bilateral auditory response. Both on the left sensors and the right sensors we observe field patterns which seem to indicate a dipolar-like activity in the temporal or central regions.
Close everything with the button [X] in the top-right corner of the Brainstorm window.
Accept to save the modifications (you deleted the "cardiac" events).- Open the "standard" average in the same way and delete the "cardiac" markers.
Repeat the same operations for Run#02:
- Open the MEG recordings for deviant and standard.
- Delete the "cardiac" markers in both files.
- Open a 2D topography and review the recordings.
- Close everything.
Interpretation
Let's display the two conditions "standard" and "deviant" side-by-side, for Run#01.
Right-click on average > MEG > Display time series.
Right-click on average > MISC > Display time series (EEG electrodes Cz and Pz)
Right-click on average > MEG > 2D Sensor cap
In the Filter tab: add a low-pass filter at 40Hz.
In the Record tab: you can set a common amplitude scale for all the figures with the button [=].
Here are the results for the standard (top) and deviant (bottom) beeps:
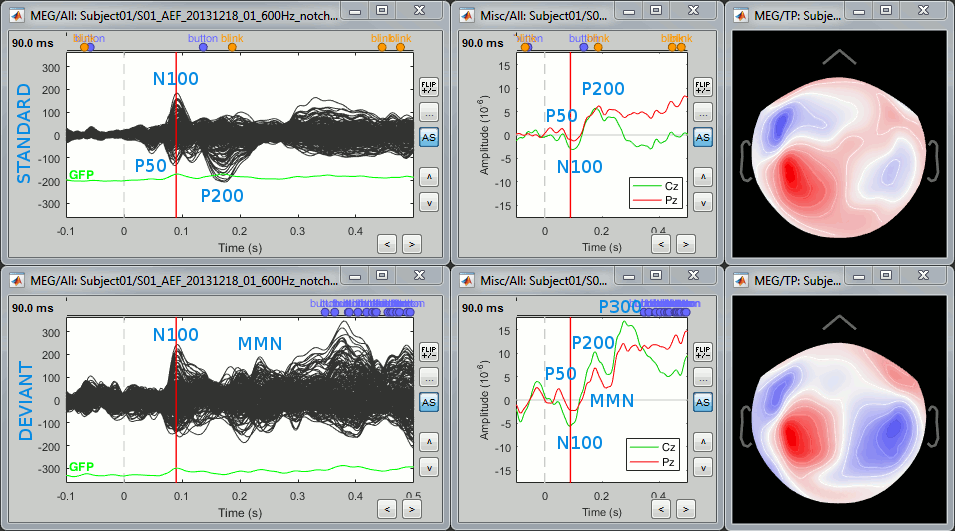
The legend in blue shows names often used in the EEG ERP literature:
P50: 50ms, bilateral auditory response in both conditions.
N100: 95ms, bilateral auditory response in both conditions.
MMN: 230ms, mismatch negativity in the deviant condition only (detection of an abnormality).
P200: 170ms, in both conditions but much stronger in the standard condition.
P300: 300ms, deviant condition only (decision making in preparation of the button press).
- Some seem to have a direct correspondance in MEG (N100) some don't (P300).
Additional quality check with the event markers:
- The time series figure for the standard condition show two unwanted events between 100ms and 200ms post-stimulus, one "blink" and one "button" response. The trials that contain them should be marked as bad and the average recomputed, because the subject is not doing the task correctly.
- We will not do it here, but keep in mind that this option "Keep all events" from the averaging process give a good summary view of the recordings and can help you identify some bad trials.
Averaging bad channels
The bad channels can be defined independently for each trial and each channel is averaged independently, therefore we can have different numbers of data points averaged for different electrodes. If we have a channel A considered good for NA trials, the corresponding channel in the average file is: sum(NA trials) / NA.
In the averaged file, a channel is consider as good if it is good in at least one trial, and considered as bad if it is bad in all the trials. The entire file is then considered as if it were computed from Nmax trials, Nmax = max(Ni, i=1..Ntrials.
This may cause some unwanted effects across channels: the SNR might be higher for some channels than others. This way of proceeding was decided to allow the conservation of the maximum amount of data. If you want to avoid this to happen: mark the channels as bad in all the trials, or report all the bad channels to the average file. This can be done easily using the database explorer, see tutorial Bad channels.
Averaging across runs
As said previously, it is usually not recommended to average recordings in sensor space across multiple acquisition runs because the subject might have moved between the sessions. Different head positions were recorded for each run, so we will reconstruct the sources separately for each each run to take into account these movements.
However, in the case of event-related studies it makes sense to start our data exploration with an average across runs, just to evaluate the quality of the evoked responses. We have seen in tutorial #4 that the subject almost didn't move between the two runs, so the error would be minimal.
Let's compute an approximate average across runs. We will run a formal average in source space later.
To run the same process again with different parameters: File > Reload last pipeline. Select:
By trial group (subject average): It will compute one average per experimental condition.
Arithmetic average + Standard error: It will save, in the same file, the average and the standard error across all the trials, illustrated in the next section.
Keep all the event markers: Do not select this option, we've already seen what it does.
The two files that are created are now saved in a new folder "Intra-subject". This is where all the results of processes involving multiple folders, within one subject, will be saved.
Standard error
If you computed the standard deviation or the standard error together with an average, it will be automatically represented in the time series figures.
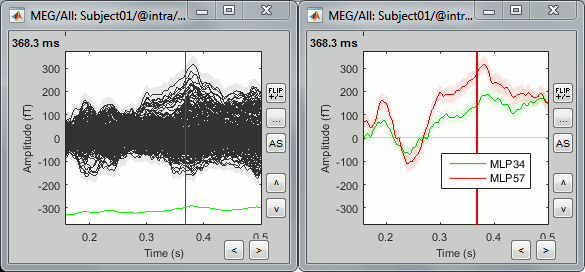
Double-click on one of the AvgStderr files to display the MEG sensors.
The light-grey area around the sensors represent the maximum standard error around the maximum and minimum values across all the sensors.Select two sensors and plot them separately (right-click > Channels > View selected, or "Enter").
The green and red areas represent, at each time point, the standard error around the signal.
Right-click on the file > File > View file contents.
The average is saved in the field F, the standard error is saved in the field Std.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
Bad channels
Number of trials
You should always be careful when comparing averages computed from different numbers of trials. In most cases, you can safely include all the trials in your averages, even in the case of imbalanced designs. However, for very low numbers of trials, having the same number of trials becomes more critical.
See the following references for more details:
Luck SJ (2010)
Is it Legitimate to Compare Conditions with Different Numbers of Trials?Thomas DG, Grice JW, Najm-Briscoe RG, Miller JW (2004)
The influence of unequal numbers of trials on comparisons of average event-related potentials
Selecting the trials
We will now compute the average responses for both the "standard" and "deviant" conditions. Two constraints have to be taken into consideration at this stage.
Using the same number of trials: As the goal in this analysis is to directly compare the two experimental conditions, we should use the same number of trials for computing the averages. We need to find the group of trials with the lowest number of good trials, and pick the same number in every group. Here we will be limited to 39 trials because of the deviant condition in Run#01. If we had multiple subjects and were planning to compute some group statistics, we should also use equal numbers of trials for all the subjects.
The instructions below show how to easily select an equal number of trials from the database:
Drag and drop all the "standard" and "deviant" trials for both runs in Process1.
Select the process "Files > Select uniform number of trials".
Select the options: By trial group (folder) and Uniformly distributed.
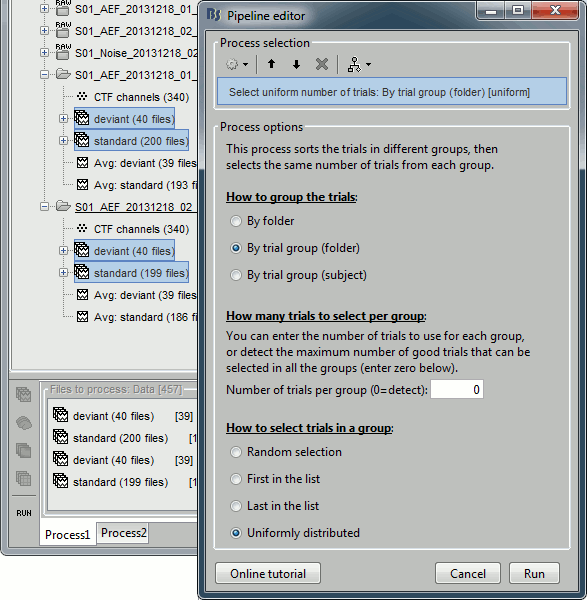
If you click on [Run], it doesn't do anything but highlighting the first selected file in the database explorer. This process just performs a file selection, it needs to be followed by another process that uses the selected files for computing something. However, you can see what was done in the process report. The reports are displayed only when an error or a warning was reported, but you can open them manually to check for additional messages. Menu File > Report viewer.
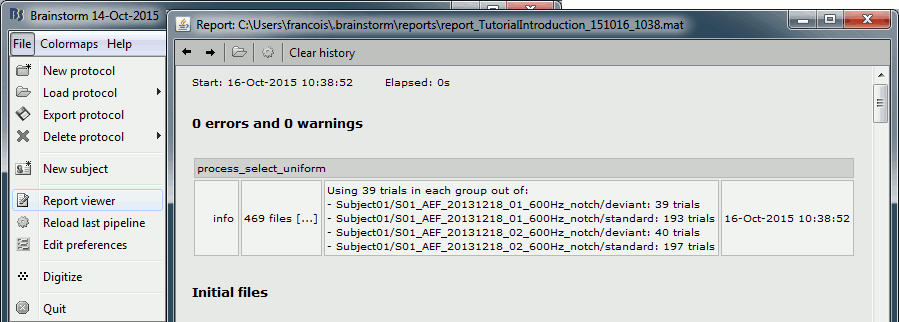
The comment in the report shows the 4 groups of trials that were identified based on the option we selected (group "by trial group and folder"), with the number of good trials per group:
Run01/deviant (39), Run01/standard (193), Run02/deviant (40), Run01/standard (197).
The process picked 39 trials in each group, uniformly distributed in the list of available trials.
Example of trial indices selected for Run01/standard: [1, 6, 11, 16, 21, 26, 31, 36, ..., 188, 193]
Process options: Select trials
Available options in the process: File > Select uniform number of trials.
By folder: Groups by subject and by folder, ignoring the trial groups.
Here, it would identify two groups, one for each acquisition run: Run01, Run02.By trial group (folder): Groups by set of trials with the same name, separately for each folder and each subject. Here it would identify four groups: Run01/deviant, Run01/standard, Run02/deviant, Run01/standard.
By trial group (subject): Groups by set of trials with the same name, for each subject. The separation in folders is ignored. Here it would identify two groups: deviant, standard.
How to select trials in a group that contains more than the requested number (Nf files, selecting only Ns):
Random selection: Select a random subset of Ns trials. Trial indices: randperm(Nf,Ns)
First in the list: Select the first Ns trials. Trial indices: 1:Ns
Last in the list: Select the last Ns trials. Trial indices: Nf-Ns+1:Nf
Uniformly distributed: Select Ns equally spaced trials. Trial indices: round(linspace(1, Nf, Ns)))