|
Size: 3882
Comment:
|
Size: 13224
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Group studies: Subject co-registration = In the typical Brainstorm workflow, you process each subject individually: you have access to the MRI of the subject, you extract the cortex envelope from it using the software of your choice (!FreeSurfer, BrainVISA, !BrainSuite or CIVET), you import the recordings and estimate the sources for each subject separately. |
= Group analysis: Subject coregistration = ''Authors: Francois Tadel'' In the typical Brainstorm workflow, you process each subject individually: you have access to the MRI of the subject, you extract the cortex envelope from it using the software of your choice (FreeSurfer, BrainVISA, BrainSuite CAT12 or CIVET), you import the recordings and estimate the sources for each subject separately. |
| Line 6: | Line 8: |
| The solution is to reproject the sources of all the subjects on the default anatomy. The reprojected versions share the same source space, the cortex from Colin27 anatomy, so they can be averaged or used to calculate any other statistics across subjects. Different methods are available in Brainstorm depending on the software you are using for the MRI segmentation. The recommended option is to use !FreeSurfer and benefit from the very efficient registration system this software offers. | The solution is to reproject the sources of all the subjects on the default anatomy. The reprojected versions share the same source space, the cortex from ICBM152 anatomy, so they can be averaged or used to calculate any other statistics across subjects. Different methods are available in Brainstorm depending on the software you are using for the MRI segmentation. The recommended option is to use FreeSurfer and benefit from the very efficient registration system this software offers. <<TableOfContents(2,2)>> |
| Line 9: | Line 13: |
| The !FreeSurfer software registers all the subjects to the FSAverage atlas using a spherical representation of each hemisphere. The hemisphere of the subject's brain are inflated to a sphere, then this sphere is deformed to match the curvature map of the equivalent sphere in the FSAverage subject. The result of this registration is saved in the !FreeSurfer output folder in '''surf/lh.sphere.reg''' and '''surf/rh.sphere.reg'''. | FreeSurfer and CAT12 register all the subjects to the FSAverage atlas using a spherical representation of the cortex. Each hemisphere of the subject's brain is inflated to a sphere, which is then deformed to match the curvature map of the equivalent sphere in the FSAverage subject. The result of this registration is saved in the FreeSurfer output folder in '''surf/lh.sphere.reg''' and '''surf/rh.sphere.reg'''. When you import the FreeSurfer anatomy folder in Brainstorm, it reads these files and save the position of the vertices in the field "'''.Reg.Sphere.Vertices'''" of the cortex, white, and mid-gray surfaces.<<BR>><<BR>> |
| Line 11: | Line 15: |
| We can illustrate this method using the protocol '''TutorialRaw''', that you created while reading the introduction tutorials. | . {{attachment:workflow_small.gif}} <<BR>><<BR>> We illustrate this method using the dataset from the tutorial [[http://neuroimage.usc.edu/brainstorm/Tutorials/MedianNerveCtf|MEG median nerve (CTF)]], but it is very easy to transpose to the protocol TutorialIntroduction you created in the introduction tutorials. |
| Line 14: | Line 20: |
| 1. Right-click on the high-resolution cortex surface (Subject01/cortex_300000V) > MRI registration > View !FreeSurfer sphere. Those two spheres represent the inflated left and the right hemispheres, registered on the FSAverage subject. The darker stripes represent the bottom of the sulci: the map is exactly the same as the one that is displayed on the cortex surface when you click on the "Sulci" button in the Surface tab. 1. Do the same for the low-resolution cortex surface (Subject01/cortex_300000V). You see the downsampled version of the same thing. 1. Now do the same for the high-resolution cortex of the default anatomy. |
1. Right-click on the high-resolution cortex surface ('''Subject01/cortex_300000V''') > MRI registration > '''View FreeSurfer sphere'''. These two spheres represent the inflated left and the right hemispheres, registered on the FSAverage subject. The darker stripes represent the bottom of the sulci: the map is exactly the same as the one that is displayed on the cortex surface when you click on the "Sulci" button in the Surface tab. 1. Do the same for the low-resolution cortex surface (Subject01/cortex_300000V). This is the downsampled version of the same thing. You can click on the "Edge" button on the surface tab to see the same triangulation as you would see when displaying the low-resolution cortex surface.<<BR>><<BR>> {{attachment:sphereSubj.gif||width="538",height="173"}} 1. Now do the same for the cortex of the '''default anatomy'''. All these spheres are registered to the FSaverage subject (the vertices of the spheres are moved to match the curvature maps of the FSAverage sphere). Hence they are also registered together: we can use them to reproject the source calculated on the individual anatomies to the default anatomy. <<BR>><<BR>> {{attachment:sphereDefault.gif||width="538",height="173"}} 1. Now switch to the functional part of the protocol. Right-click on the average sources for condition Left > '''Cortical activations > Display on spheres'''. Double-click on the file to display the regular brain at the same time. Change the current time to a meaningful event (35ms for instance).<<BR>><<BR>> {{attachment:sourcesSubj.gif||width="441",height="164"}} 1. To project this source file to the default anatomy ICBM152, right-click on the Left/Average/MN source file > '''Project sources > Default anatomy > cortex_15000V'''. The source values are re-interpolated from the registered spheres of Subject01 to the spheres of ICBM152 using the Shepard’s method (weighted combination of a few nearest neighbors) <<BR>><<BR>> {{attachment:projectRaw.gif||width="402",height="230"}} 1. It creates a new subject "Group analysis" to store the projected source maps:<<BR>><<BR>> {{attachment:projectTreeRaw.gif||width="402",height="211"}} 1. Double-click on this new file, and display it on the registered spheres as well:<<BR>><<BR>> {{attachment:sourcesDefault.gif||width="440",height="164"}} |
| Line 18: | Line 28: |
| The best available option is selected automatically | == Projection on high-resolution cortex == You can also use the same method to re-project the sources on the high-resolution cortex, to produce nicer figures. The process is the same: right-click on Left/Average/MN source file > '''Project sources > Subject01 > cortex_300000V'''. It creates a new file in the same condition. |
| Line 20: | Line 31: |
| == Those menus don't show up == If you have started your analysis before those options were made available in Brainstorm |
. {{attachment:projectTreeHires.gif}} {{attachment:sourcesHires.gif||width="440",height="164"}} Important notes: * You probably don't need 300,000 vertices on the cortex to get figures nice enough for your publications. You can downsample the high-resolution to something around 90,000 vertices, and then re-project the sources on this mid-resolution brain. * To get a smoother display instead of the all these little dots due to the Shepards interpolation, you can run the process Sources > Spatial smoothing on the high-resolution cortical maps. == It doesn't work == If you have started your analysis before these options were made available in Brainstorm you would probably not be able to apply these operations to your own data. * You don't see the menus described above: update Brainstorm. * You get the error "There is no registered sphere available for this surface" for the subject cortex: * Re-import the anatomy of the subject from the FreeSurfer folder after making sure that the files surf/?h.sphere.reg are not missing. Then delete all the head models and sources results, and re-calculate them. * Warning: Don't do this if you already have a lot of results for this subject, it would probably make all the source results unusable. * You get this error for the default anatomy: * You have to update the default anatomy for this protocol. Right-click on the folder (Default anatomy) > Use template > ICBM152 * Warning: Don't do this if you already have projected all your results on the default anatomy, or if you have some subjects that use the default anatomy for the source estimation. It would make all the results unusable for sure. * When you use the menu "Project sources", you see that it's using the second method described below (you see figures with these smoothed red and gray surfaces): it means that either the subject or the default anatomy doesn't have the registered spheres saved in its structure. You should also observe one or the other of the above error, fix accordingly. == Registration with BrainSuite == BrainSuite also provides an accurate registration method to the Brainstorm anatomy templates (Colin27, ICBM152). The registration procedure works in the same way as the FreeSurfer-based registration illustrated in this tutorial. To use this registration, you need to use a template anatomy processed with BrainSuite. <<BR>> Right-click on ''Default anatomy'' > Use template > '''Colin27_BrainSuite''' or '''ICBM152_BrainSuite'''. . {{attachment:coreg.gif||width="492",height="293"}} == Warped brains == It is important to note that pseudo-individual anatomies that were generated by deforming the ICBM152 brain using the digitized head points '''do not need''' to be re-projected on the default anatomy. If you have two subjects for which you generated pseudo anatomies using the popup menu ''Digitized head points > Warp > Deform default anatomy to fit these points'', you can directly use their source maps for computing any statistics or difference. You don't need to use this menu "Project sources". {{attachment:warp.jpg}} == Volume source models == Source files using volume head models contain a list of grid points that are not necessarily positioned on the cortex surface. We cannot use the surface-based atlases from FreeSurfer or BrainSuite. Instead, we rely on the [[http://neuroimage.usc.edu/brainstorm/CoordinateSystems#MNI_coordinates|MNI transformation]] computed using SPM functions. The procedure to project these source files on a template is different: You need to define a grid at the template level, then project this grid in the subject space and estimate the sources for this grid. * Create a "Group analysis" entry (if not already existing): <<BR>><<BR>> {{attachment:volume_group.gif||width="270",height="136"}} * Generate a template grid in the group analysis folder: <<BR>><<BR>> {{attachment:volume_generate.gif||width="510",height="190"}} * Compute a head model for each subject in your database: Use the option "MRI volume" and then<<BR>>"Use template grid for group analysis".<<BR>><<BR>> {{attachment:volume_headmodel.gif||width="621",height="355"}} * Estimate the sources for each subject, then project them on the template: <<BR>><<BR>> {{attachment:volume_project.gif||width="458",height="172"}} * The two source files you obtain (original and projected) contain exactly the same information, with the same number of sources, but they are displayed in the different spaces. If you display the two files side by side, the original source file is displayed on the subject MRI (left) while the projected file is displayed on the template MRI from the default anatomy folder (right). <<BR>><<BR>> {{attachment:volume_display.gif||width="457",height="239"}} * If you project source files from multiple subjects in the same way, you will be able to average them or compute statistics across subjects after this projection. == Mixed head models == The mixed head models introduced in the tutorial [[http://neuroimage.usc.edu/brainstorm/Tutorials/DeepAtlas|Deep cerebral structures]] are not suitable for group analysis. We do not have any accurate solution for projecting the individual source space to the template space. Use volume head models instead. |
| Line 24: | Line 80: |
| If you have not processed your MRI files with !FreeSurfer, you still have an option available to re-project your subject's source maps on the default anatomy. We can illustrate this method using the protocol '''TutorialCTF''', that you created while reading the introduction tutorials. | If you did not process your MRI files with FreeSurfer, or if you are working with an older database, you have another option available to re-project your subject's source maps on the default anatomy. The results are much less reliable, but the method is quite robust and can be used for all sorts of purposes. We can illustrate this method using the protocol '''TutorialCTF''', that you created while reading the introduction tutorials. |
| Line 26: | Line 82: |
| == Warped brains == It is important to note that pseudo-individual anatomies that were generated by deforming the Colin27 brain using the digitized head points do not need to be re-projected on the default anatomy. If you have two subjects for which you generated pseudo anatomies using the popup menu Digitized head points > Warp > Deform default anatomy to fit these points, you can directly use their source maps for computing any statistics or difference. You don't need to use this menu "Project sources". |
Running this method is done exactly in the same way as with the FreeSurfer spheres, using the "Project sources" menu. There is no additional option, Brainstorm would just automatically choose the best method available for the source projection. If the registered spheres are available in the field ".Reg.Sphere.Vertices", it uses them, if not it uses this method. |
| Line 29: | Line 84: |
| == Feedback == <<EmbedContent(http://neuroimage.usc.edu/brainstorm3_register/get_feedback.php?Tutorials/TutFirstSteps)>> |
1. Right-click on Subject01/Right/MN > Project sources > Default anatomy > Cortex_15000V<<BR>><<BR>> {{attachment:projectCTF.gif||width="425",height="376"}} 1. The source cortex (Subject01) and the destination cortex (ICBM152) are split in two hemispheres, and then for each hemisphere: * Alignment along the anterior commissure/posterior commissure axis * Smooth intensely to keep only the main featuresof the surfaces * Deformation of the individual surface to match the ICBM152 surface with an iterative closest point algorithm (ICP). The following figures show the left hemispheres before and after the ICP deformation:<<BR>><<BR>> {{attachment:projectBefore.gif||width="237px",height="162px"}} {{attachment:projectAfter.gif||width="235px",height="161px"}} 1. Interpolation of the source amplitudes from the subject surface to the ICBM152 surface using the Shepard’s method (weighted combination of a few nearest neighbors) 1. The interpolation matrix subject->Colin is computed only once and then saved in the subject's cortex surface file, in the field "tess2tess_interp" 1. You can see that a new subject and a new condition were created in the database, that contain the sources from Subject01/Right/ERF projected on the default anatomy:<<BR>><<BR>> {{attachment:projectTree.gif||width="364",height="272"}} 1. Display at the same time the original source file and the projected one to see how it looks like (left: Subject01, right: Colin)<<BR>><<BR>> {{attachment:projectCompare.gif||width="285",height="144"}} 1. Delete the "Group analysis" subject. == Additional discussions on the forum == * Spatial smoothing of sources: http://neuroimage.usc.edu/forums/showthread.php?1409 <<EmbedContent(http://neuroimage.usc.edu/bst/get_feedback.php?Tutorials/CoregisterSubjects)>> |
Group analysis: Subject coregistration
Authors: Francois Tadel
In the typical Brainstorm workflow, you process each subject individually: you have access to the MRI of the subject, you extract the cortex envelope from it using the software of your choice (FreeSurfer, BrainVISA, BrainSuite CAT12 or CIVET), you import the recordings and estimate the sources for each subject separately.
After importing and pre-processing all the individual information, you will probably be interested in calculating some group statistics, to extract the significant features across all the subjects in your study. in order to do this at the source level, all the source files must be using the same support cortex source (= the same source space). The problem you typically face is that the shape and number of vertices of the cortex surface varies across subject: one source index doesn't mean the same thing for all the subjects, hence it's impossible to compare them directly.
The solution is to reproject the sources of all the subjects on the default anatomy. The reprojected versions share the same source space, the cortex from ICBM152 anatomy, so they can be averaged or used to calculate any other statistics across subjects. Different methods are available in Brainstorm depending on the software you are using for the MRI segmentation. The recommended option is to use FreeSurfer and benefit from the very efficient registration system this software offers.
Contents
Using FreeSurfer registered spheres
FreeSurfer and CAT12 register all the subjects to the FSAverage atlas using a spherical representation of the cortex. Each hemisphere of the subject's brain is inflated to a sphere, which is then deformed to match the curvature map of the equivalent sphere in the FSAverage subject. The result of this registration is saved in the FreeSurfer output folder in surf/lh.sphere.reg and surf/rh.sphere.reg. When you import the FreeSurfer anatomy folder in Brainstorm, it reads these files and save the position of the vertices in the field ".Reg.Sphere.Vertices" of the cortex, white, and mid-gray surfaces.
We illustrate this method using the dataset from the tutorial MEG median nerve (CTF), but it is very easy to transpose to the protocol TutorialIntroduction you created in the introduction tutorials.
Select the protocol TutorialRaw, go the anatomy view
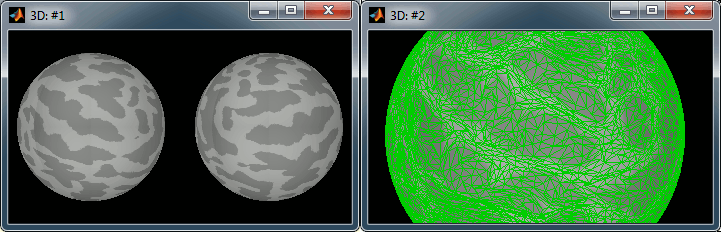
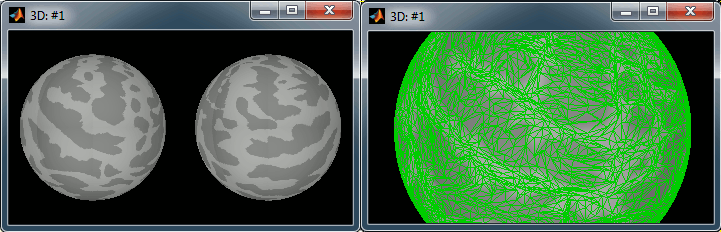
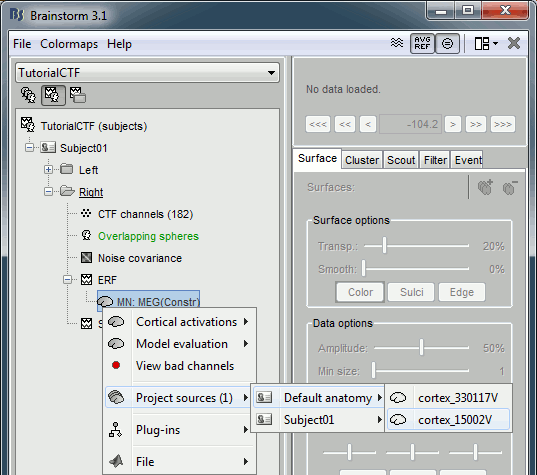
Right-click on the high-resolution cortex surface (Subject01/cortex_300000V) > MRI registration > View FreeSurfer sphere. These two spheres represent the inflated left and the right hemispheres, registered on the FSAverage subject. The darker stripes represent the bottom of the sulci: the map is exactly the same as the one that is displayed on the cortex surface when you click on the "Sulci" button in the Surface tab.
Do the same for the low-resolution cortex surface (Subject01/cortex_300000V). This is the downsampled version of the same thing. You can click on the "Edge" button on the surface tab to see the same triangulation as you would see when displaying the low-resolution cortex surface.
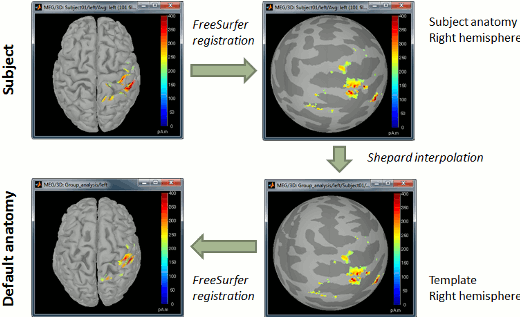
Now do the same for the cortex of the default anatomy. All these spheres are registered to the FSaverage subject (the vertices of the spheres are moved to match the curvature maps of the FSAverage sphere). Hence they are also registered together: we can use them to reproject the source calculated on the individual anatomies to the default anatomy.
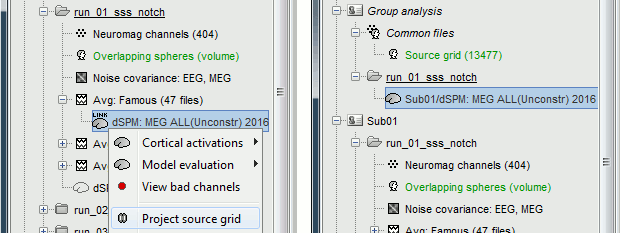
Now switch to the functional part of the protocol. Right-click on the average sources for condition Left > Cortical activations > Display on spheres. Double-click on the file to display the regular brain at the same time. Change the current time to a meaningful event (35ms for instance).
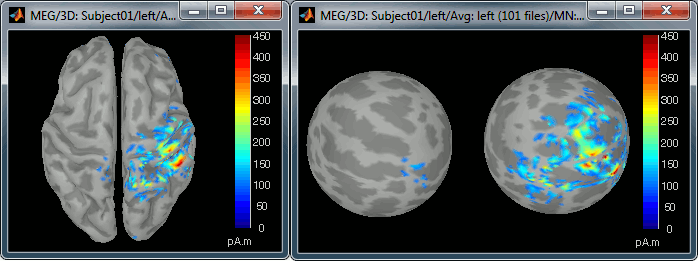
To project this source file to the default anatomy ICBM152, right-click on the Left/Average/MN source file > Project sources > Default anatomy > cortex_15000V. The source values are re-interpolated from the registered spheres of Subject01 to the spheres of ICBM152 using the Shepard’s method (weighted combination of a few nearest neighbors)
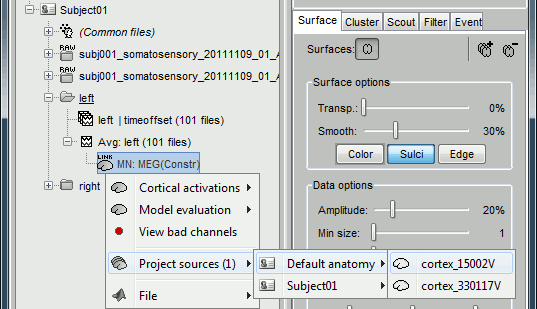
It creates a new subject "Group analysis" to store the projected source maps:
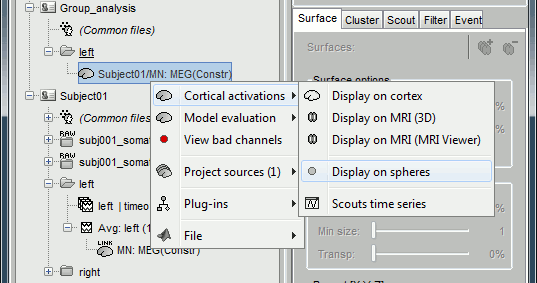
Double-click on this new file, and display it on the registered spheres as well:

Projection on high-resolution cortex
You can also use the same method to re-project the sources on the high-resolution cortex, to produce nicer figures. The process is the same: right-click on Left/Average/MN source file > Project sources > Subject01 > cortex_300000V. It creates a new file in the same condition.
Important notes:
- You probably don't need 300,000 vertices on the cortex to get figures nice enough for your publications. You can downsample the high-resolution to something around 90,000 vertices, and then re-project the sources on this mid-resolution brain.
To get a smoother display instead of the all these little dots due to the Shepards interpolation, you can run the process Sources > Spatial smoothing on the high-resolution cortical maps.
It doesn't work
If you have started your analysis before these options were made available in Brainstorm you would probably not be able to apply these operations to your own data.
- You don't see the menus described above: update Brainstorm.
- You get the error "There is no registered sphere available for this surface" for the subject cortex:
Re-import the anatomy of the subject from the FreeSurfer folder after making sure that the files surf/?h.sphere.reg are not missing. Then delete all the head models and sources results, and re-calculate them.
- Warning: Don't do this if you already have a lot of results for this subject, it would probably make all the source results unusable.
- You get this error for the default anatomy:
You have to update the default anatomy for this protocol. Right-click on the folder (Default anatomy) > Use template > ICBM152
- Warning: Don't do this if you already have projected all your results on the default anatomy, or if you have some subjects that use the default anatomy for the source estimation. It would make all the results unusable for sure.
- When you use the menu "Project sources", you see that it's using the second method described below (you see figures with these smoothed red and gray surfaces): it means that either the subject or the default anatomy doesn't have the registered spheres saved in its structure. You should also observe one or the other of the above error, fix accordingly.
Registration with BrainSuite
BrainSuite also provides an accurate registration method to the Brainstorm anatomy templates (Colin27, ICBM152). The registration procedure works in the same way as the FreeSurfer-based registration illustrated in this tutorial.
To use this registration, you need to use a template anatomy processed with BrainSuite.
Right-click on Default anatomy > Use template > Colin27_BrainSuite or ICBM152_BrainSuite.
Warped brains
It is important to note that pseudo-individual anatomies that were generated by deforming the ICBM152 brain using the digitized head points do not need to be re-projected on the default anatomy.
If you have two subjects for which you generated pseudo anatomies using the popup menu Digitized head points > Warp > Deform default anatomy to fit these points, you can directly use their source maps for computing any statistics or difference. You don't need to use this menu "Project sources".
Volume source models
Source files using volume head models contain a list of grid points that are not necessarily positioned on the cortex surface. We cannot use the surface-based atlases from FreeSurfer or BrainSuite. Instead, we rely on the MNI transformation computed using SPM functions.
The procedure to project these source files on a template is different: You need to define a grid at the template level, then project this grid in the subject space and estimate the sources for this grid.
Create a "Group analysis" entry (if not already existing):
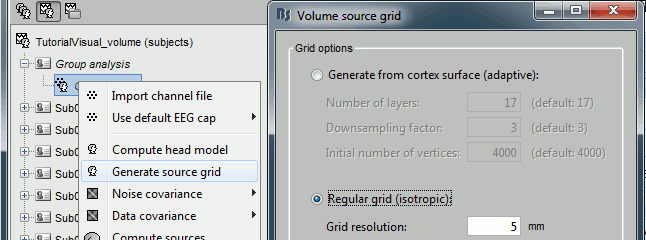
Generate a template grid in the group analysis folder:
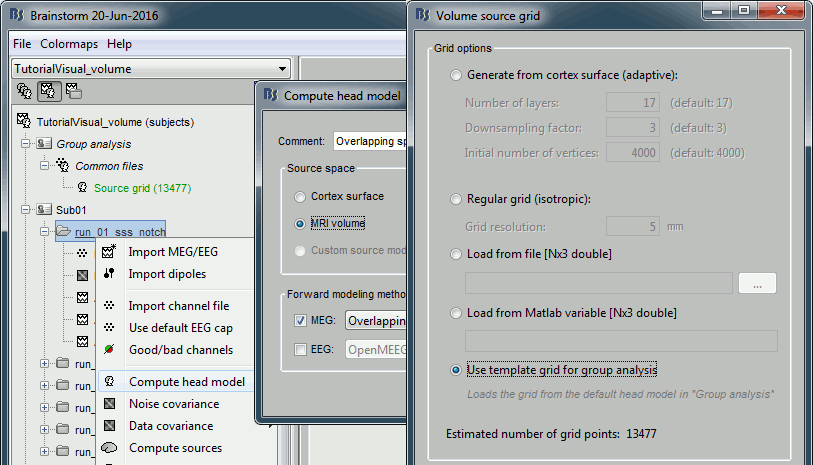
Compute a head model for each subject in your database: Use the option "MRI volume" and then
"Use template grid for group analysis".
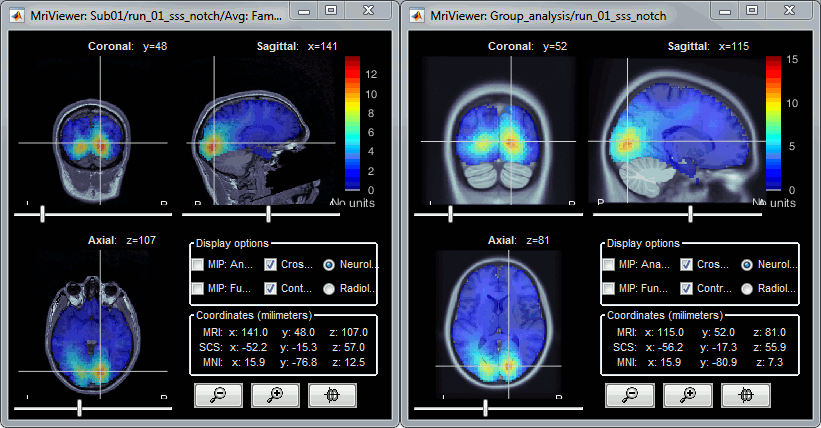
Estimate the sources for each subject, then project them on the template:
The two source files you obtain (original and projected) contain exactly the same information, with the same number of sources, but they are displayed in the different spaces. If you display the two files side by side, the original source file is displayed on the subject MRI (left) while the projected file is displayed on the template MRI from the default anatomy folder (right).
- If you project source files from multiple subjects in the same way, you will be able to average them or compute statistics across subjects after this projection.
Mixed head models
The mixed head models introduced in the tutorial Deep cerebral structures are not suitable for group analysis. We do not have any accurate solution for projecting the individual source space to the template space. Use volume head models instead.
A cheap alternative
If you did not process your MRI files with FreeSurfer, or if you are working with an older database, you have another option available to re-project your subject's source maps on the default anatomy. The results are much less reliable, but the method is quite robust and can be used for all sorts of purposes. We can illustrate this method using the protocol TutorialCTF, that you created while reading the introduction tutorials.
Running this method is done exactly in the same way as with the FreeSurfer spheres, using the "Project sources" menu. There is no additional option, Brainstorm would just automatically choose the best method available for the source projection. If the registered spheres are available in the field ".Reg.Sphere.Vertices", it uses them, if not it uses this method.
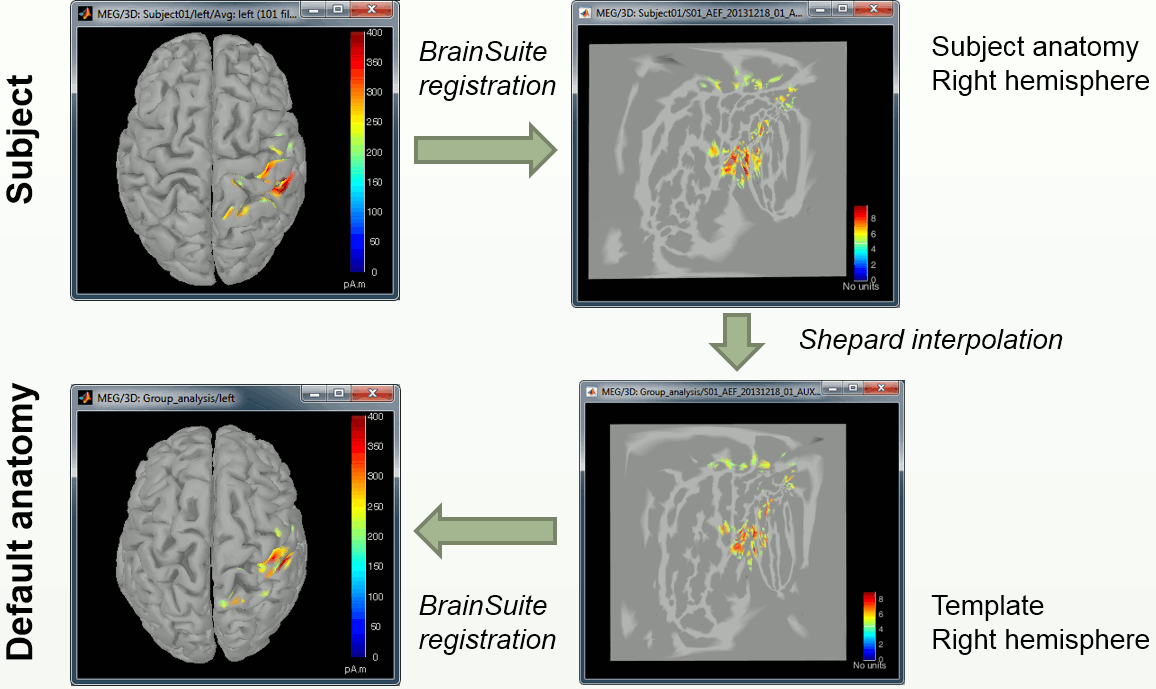
Right-click on Subject01/Right/MN > Project sources > Default anatomy > Cortex_15000V
- The source cortex (Subject01) and the destination cortex (ICBM152) are split in two hemispheres, and then for each hemisphere:
- Alignment along the anterior commissure/posterior commissure axis
- Smooth intensely to keep only the main featuresof the surfaces
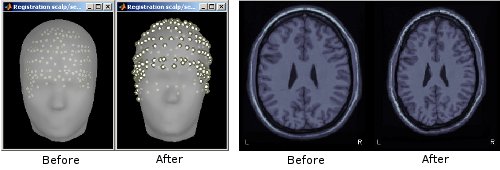
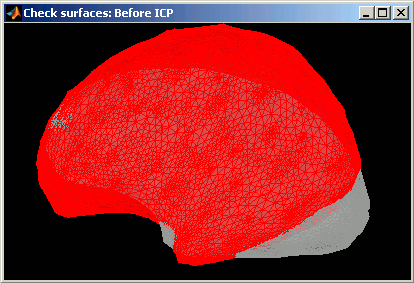
Deformation of the individual surface to match the ICBM152 surface with an iterative closest point algorithm (ICP). The following figures show the left hemispheres before and after the ICP deformation:
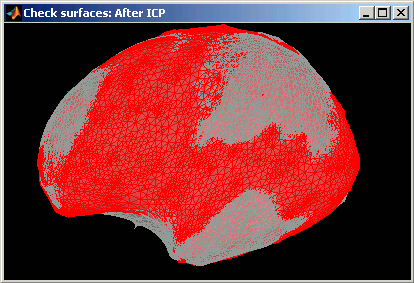
- Interpolation of the source amplitudes from the subject surface to the ICBM152 surface using the Shepard’s method (weighted combination of a few nearest neighbors)
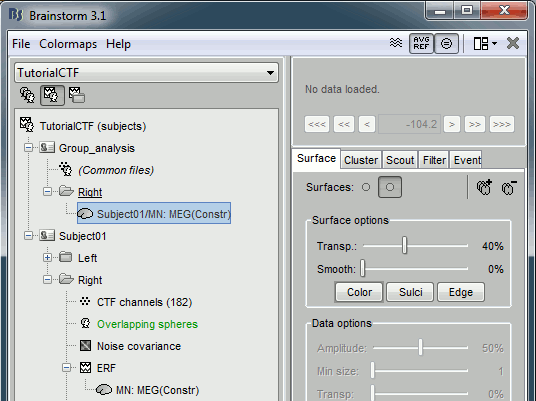
The interpolation matrix subject->Colin is computed only once and then saved in the subject's cortex surface file, in the field "tess2tess_interp"
You can see that a new subject and a new condition were created in the database, that contain the sources from Subject01/Right/ERF projected on the default anatomy:
Display at the same time the original source file and the projected one to see how it looks like (left: Subject01, right: Colin)
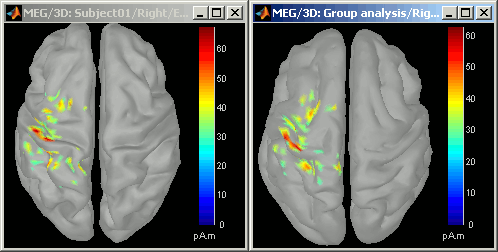
- Delete the "Group analysis" subject.
Additional discussions on the forum
Spatial smoothing of sources: http://neuroimage.usc.edu/forums/showthread.php?1409