|
Size: 21817
Comment:
|
Size: 13613
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| ''Authors: Francois Tadel, Sylvain Baillet'' | ''Authors: Francois Tadel, Elizabeth Bock, Sylvain Baillet'' |
| Line 5: | Line 5: |
== How to read these tutorials == The goal of these introduction tutorials is to guide you through most of the features of the software. All the pages use the same example dataset. '''The results of one section are most of the time used in the following section''', so read these pages in the correct order. <<TAG(Advanced)>> Some pages may contain too many details for your level of interest or competence. The sections marked as '''[Advanced]''' are not required for you to follow the tutorials until the end. You can skip them the first time you go through the documentation. You will be able to get back to the theory later if you need. '''Please follow first these tutorials with the data we provide'''. This way you will be able to focus on learning how to use the software. It is better to start with some data that is easy to analyze. After going through all the tutorials, you should be comfortable enough with the software to start analyzing your own recordings. If you are interested only in''' EEG or intra-cranial recordings''', don't think that an MEG-based tutorial is not adapted for you. Most of the practical aspects of the data manipulation is very similar in EEG and MEG. Start by reading these introduction tutorials, then go through the tutorial "EEG and Epilepsy" to get some more details about the processing steps that are specific for EEG. == Presentation of the experiment == All the introduction tutorials are based on a simple auditory oddball experiment: * One subject, two acquisition runs of 6 minutes each * Subject stimulated binaurally with intra-aural earphones * Each run contains 200 regular beeps and 40 easy deviant beeps * Recordings with a CTF MEG system with 275 axial gradiometers * Anatomy of the subject: 1.5T MRI, processed with FreeSurfer 5.3 * More details will be given about this dataset along the process. * Full dataset description available on this page: [[DatasetIntroduction|Introduction dataset]] |
|
| Line 50: | Line 72: |
| 1. If you haven't read in the installation instructions, do it now: [[Installation]]. | 1. If you haven't read the installation instructions, do it now: [[Installation]]. |
| Line 52: | Line 74: |
| 1. Start Matlab, go to brainstorm3 directory, and type "brainstorm" in Matlab command window.<<BR>> | 1. Start Brainstorm from Matlab or with the compiled executable.<<BR>> |
| Line 73: | Line 95: |
| The Brainstorm window described below is designed to remain on one side of the screen. All the space of the desktop that is not covered by this window will be used for opening other figures. Do not try to maximize this window, or the automatic management of the data figures might not work correctly. Keep it on one side of your screen, just large enough so you can read the file names in the database explorer. |
|
| Line 83: | Line 109: |
| * Your Brainstorm database is a collection of protocols. | |
| Line 86: | Line 113: |
| * Anatomy: Includes at least an MRI volume and some surfaces extracted from the MRI. * Functional data: Everything that is related with the MEG/EEG acquisition. |
* '''Anatomy''': Includes at least an MRI volume and some surfaces extracted from the MRI. * '''Functional data''': Everything that is related with the MEG/EEG acquisition. |
| Line 92: | Line 119: |
| * The current structure of the database does not allow to have more that one level of sub-folders for each subject. It is not possible to organize the files by session AND by condition.<<BR>><<BR>> {{attachment:db.gif}} | * The current structure of the database does not allow more than one level of sub-folders for each subject. It is not possible to organize the files by session AND by condition.<<BR>><<BR>> {{attachment:db.gif}} |
| Line 100: | Line 127: |
| * Most of the files you see in the database explorer in Brainstorm correspond to files on the hard drive, but there is no one-to-one correspondance. There is extra information stored in each directory, to save properties, comments, default data, links between different items, etc. For this reason, you should not try to manipulate directly the files in the Brainstorm database directory. * The structure of the database is saved in the user preferences, so when you start the program or change protocol, there is no need to read again all the files on the hard drive (which may take some time). * If Brainstorm or Matlab crashes before the database structure is correctly saved, the files that are displayed in the Brainstorm database explorer may differ from what is actually on the disk. When this happens, you can force Brainstorm to rebuild the structure from the files on the hard drive: right-click on the folder to udpate > Reload. |
* Most of the files you see in the database explorer in Brainstorm correspond to files on the hard drive, but there is no one-to-one correspondence. There is extra information stored in each directory, to save properties, comments, default data, links between different items, etc. This is one of the reasons for which you should not try to manipulate directly the files in the Brainstorm database directory. * The structure of the database is saved in the user preferences, so when you start the program or change protocol, there is no need to read again all the files on the hard drive. * If Brainstorm or Matlab crashes before the database structure is correctly saved, the files that are displayed in the Brainstorm database explorer may differ from what is actually on the disk. When this happens, you can force Brainstorm to rebuild the structure from the files on the hard drive: right-click on a folder > '''Reload'''. |
| Line 104: | Line 131: |
| == Create first protocol == 1. Click on the File menu, and select, and select ''New protocol''.<<BR>><<BR>> {{attachment:menuFile.gif}} 1. Edit the protocol name and enter: "TutorialFirstSteps". It will automatically update the paths (''Anatomy path'' and ''Datasets path''). |
== Create your first protocol == 1. Menu File > New protocol.<<BR>><<BR>> {{attachment:menuFile.gif}} 1. Edit the protocol name and enter: "'''TutorialIntroduction'''".<<BR>>It will automatically update the anatomy and datasets paths. |
| Line 108: | Line 135: |
| 1. ''Default properties for the subjects'': * These are the default settings that are used when creating new subjects. It is then possible to override those settings for each subject individually. |
1. Default properties for the subjects: These are the default settings that are used when creating new subjects. It is then possible to override these settings for each subject individually. * '''Default anatomy''': (MRI and surfaces) * '''No, use individual anatomy''':<<BR>>Select when you have individual MRI scans for all the participants of your study. |
| Line 111: | Line 139: |
| * Our objective is to create one subject that uses the default anatomy Colin27, so select the option "''Yes, use protocol's default anatomy''". | * '''Yes, use default anatomy''':<<BR>>Select when you do not have individual scans for the participants, and you would like to use one of the anatomy templates available in Brainstorm. |
| Line 113: | Line 141: |
| * The option ''Default channel file'' is a bit more complicated to understand. It defines at what level we want to define the sensors information (position, orientation, name...) in the database. To make you data management more efficient, you can in some cases use the same ''channel file'' for all the subjects, or for all the folders within one subject. For more information on this option, click on the '''Help button'''. | * '''Default channel file''': (Sensors names and positions) * '''No, use one channel file per condition/run''':<<BR>>Select this if you may have different head positions for one subject. This is usually not the case in EEG, where the electrodes stay in place for all the experiment. In MEG, this is a common setting: one recording session is split in multiple acquisition runs, and the position of the subject's head in the MEG might be different between two runs. |
| Line 115: | Line 144: |
| * In this specific case, this option is not relevant because we are not going to import any recordings. | * '''Yes, use one channel file per subject''':<<BR>>This is a common setting for EEG. The electrodes are in the same position for all the files recorded on one subject. |
| Line 117: | Line 146: |
| 1. Once you get something like the following figure, click on ''Create''.<<BR>><<BR>> {{attachment:createNewProtocol.gif}} | * '''Yes, use only one global channel file''':<<BR>>This is never a recommended setting. It could be used in the case of an EEG study where you use only standard EEG positions on a standard anatomy, but only if you are not doing any advanced source reconstruction. If you share the position of the electrodes between all the subjects, it will also share the source models, which are dependent on the quality of the recordings for each subject. This is complicated to understand at this level, it will make more sense later in the tutorials. 1. In the context of this study, we are going to use the following settings:<<BR>> * '''No, use individual anatomy''': Because we have access to a T1 MRI scan of the subject. * '''No, use one channel file per condition/run''': The typical MEG setup. 1. Click on [Create].<<BR>><<BR>> {{attachment:createNewProtocol.gif}} |
| Line 122: | Line 157: |
| * You can switch between anatomy and functional data with the first three buttons in the toolbar. There are no subjects in the database yet, so the ''Functional data'' views are empty. However, in the ''Anatomy ''view, there is a ''(Default anatomy)'' node; it contains the MRI and the surfaces that can be used by default for the subjects without individual anatomy. | * You can switch between anatomy and functional data with the three buttons just above the database explorer. Read the tooltips of the buttons to see which one does what. * In the ''Anatomy ''view, there is a ''Default anatomy'' node. It contains the Colin27 anatomy, distributed by the Montreal Neurological Institute (MNI), which is one of the template anatomy folders that are distributed with Brainstorm. * The ''Default anatomy'' node contains the MRI and the surfaces that are used for the subjects without an individual anatomy, or for registering multiple subjects to the same anatomy for group analysis. * There are no subjects in the database yet, so the ''Functional data'' views are empty. |
| Line 124: | Line 162: |
| * The default anatomy used in Brainstorm is based on the ''Colin27 ''MRI volume provided by the Montreal Neurological Institute (MNI): an average of 27 T1 scans of Colin Holmes brain ([[http://www.bic.mni.mcgill.ca/~louis/stx_history.html|more info]]). * The cortex surfaces here were extracted from this average MRI with FreeSurfer. * The inner and outer skull surfaces were generated with Brainstorm. * Display the ''Anatomy ''view, and click on the small "+" to expand the contents of the ''(Default anatomy)'' node. There is one MRI and several surfaces, identified by different icons. * Everything you can do with a node (anatomy files, subjects, protocol) is accessible by right-clicking on it. Take a few minutes to explore all the popup menus by yourself. Don't be afraid of clicking everywhere, at this point you cannot damage anything. |
* Everything you can do with an object in the database explorer (anatomy files, subjects, protocol) is accessible by right-clicking on it. <<BR>><<BR>> |
| Line 135: | Line 165: |
== Changing the temporary folder == If the amount of storage space you have for your user folder is limited (less than 10Gb), you may have to change the temporary folder used by Brainstorm. If you work on a centralized network where all the computers are sharing the same resources, the system admins may impose limited disk quotas to all users and encourage them to use local hard drives instead of the limited and shared user folder. The default temporary folder of Brainstorm is located in your user folder ($HOME/.brainstorm/tmp/), so when you import recordings or calculate large models, it may quickly fill up your limited quota and at some point block your user account. To prevent this, select the menu '''File > Edit preferences''' and set the temporary directory to a folder that is local to your computer, in which you won't suffer from any storage limitation. . {{attachment:preferences.gif||height="293",width="503"}} |
|
| Line 143: | Line 180: |
| * Do not try to maximize the Brainstorm window: keep it small on one side of your screen. | |
| Line 144: | Line 182: |
| == Fiducials selection (MRI Viewer) == A few more details about the MRI and the fiducials: Right-click on the MRI file > Edit MRI... |
<<HTML(<!-- END-PAGE -->)>> |
| Line 147: | Line 184: |
| . {{attachment:mriViewer.gif}} * Navigate in the volume by clicking anywhere in the slices, or by using the sliders below each image. * Once you have clicked on any orientation, you can use the '''mouse wheel''' to move to the next/previous slice in the same orientation. * '''Zoom '''in and out with the magnifying glass buttons at the bottom of the figure * There are three small buttons above each image. They may be useful when you import your own MR volumes, if they are not oriented in the way Brainstorm wants them (e.g. you see the ''Sagittal ''title on top of the coronal view; or the small white "''P''", which is supposed to indicate the posterior part of the head, points the nose). Leave your mouse a few seconds over any button to read its description tooltip. Click on them and try to understand what they are doing. * Image contrast can be changed with the mouse: click and hold the right button on one image, and move your mouse up and down (this operation will be later referred as ''right-click+move''). * Right-clicking in any view shows a popup menu that will be described in the following tutorials. * You can define six points on the MRI: * Three to define the Subject Coordinate System (SCS):<<BR>>Nasion (NAS), Left pre-auricular point (LPA), Right pre-auricular point (RPA) * Three to define the Normalized coordinate system (NCS):<<BR>>Anterior commissure (AC), Posterior commissure (PC), and any Interhemispheric point (IH) * More information on how to define the fiducial points on the CoordinateSystems page. * To define a point, position the three views at the point your want to define (pointed by the 3 crosses), and click on the ''Set ''button. Later you can click on ''View ''to move the views to this point again. * You can see that all the points are already defined on this MRI, but you should always check their positions. For many reasons, each research group has a different way to define those points. So you may not position the ears or the nasion exactly the same way as we do. Do not hesitate to move the fiducials to better fit your needs. * The ''Display options'' panel allows to hide some of the components. If the ''MIP/Anatomy ''checkbox is selected, the Maximum Intensity Power is displayed instead of the regular slices: At each point, the value displayed is the maximum value across all the slices. * The ''Coordinates ''panel shows the (x,y,z) coordinates in the coordinate systems managed by Brainstorm: MRI and SCS. * Once you're done, click on ''Cancel ''(unless you really want to save all your experiments). == MRI visualization == There are many different ways to display the MR volumes. Right click on the ''T1-MRI'' in ''Default anatomy'', menu ''Display''. Just click everywhere and try all the options by yourself, it is the best way to learn.<<BR>><<BR>> {{attachment:popupMri.gif}} ==== MRI Viewer ==== Already introduced in this tutorial<<BR>>It is the default visualization mode; when you double-click on the MRI file, it brings up the MRI Viewer. ==== Axial / coronal / sagittal slices ==== {{attachment:mriSlicesContact.gif||height="231",width="210"}} * '''Zoom''': mouse wheel (or two finger-move on a MacBook pad) * '''Move '''in zoomed image: click + move * '''Adjust contrast''': right click + move up/down * Colormap selection in menu * Save as image in menu ==== 3D orthogonal slices ==== ||<tablewidth="200px"> {{attachment:mriSlices3d.gif||height="229",width="292"}} || {{attachment:menuShortcut.gif}} || * Simple mouse operations * '''Rotate''': click + move * '''Zoom''': mouse wheel (two finger-move on a MacBook pad) * '''Move object''': left+right click + move * '''Move MRI slices''': right click + move along direction of the slice<<BR>>(or use the ''Resect ''panel in the ''Surface ''tab) * '''Colormap contrast/brightness''': click on the colorbar, and move up/down (brightness) or left/right (contrast) * '''Reset view''': double click * '''Reset colormap''': double-click on the colorbar * Popup operations (right-click on the figure) * '''Colormap''': Full ''Anatomy ''colormap edition (detailed in next tutorial) * '''MRI Display''': For now, contains only the MIP option (Maximum Intensity Power). If checked, for each orientation: displays the maximum along all the slices instead of the proper slice. * '''Get coordinates''': Pick a point in any 3D view and get its coordinates * '''Snapshots''': save images or movies from this figure * '''Figure''': change background color, display axes and menus for an advanced figure editing and management using Matlab tools. * '''Views''': configure camera position with predefined settings. * Keyboard shortcuts: * Notice the indications in the right part of the popup menu (''CTRL+A'' in front of ''Figures>Axes'', ''"1"'' in front of ''Views>Left''), they represent the keyboard shortcut for each menu. * '''Views shortcuts (0,1,2...9 and "=")''': Remember them, they will be very useful when exploring the cortical sources. It is much faster to press a key to switch from left to right hemisphere, than having to rotate the brain with the mouse. * Surfaces tab (in the Brainstorm main window): * This panel is primarily dedicated to the surfaces display, but some controls can also be useful for the 3D MRI view. * ''Transparency ''slider * ''Smooth ''slider: it changes the threshold applied to the MRI slices. If you set it zero, you will see the full slices, as extracted from the volume. * ''Resect ''panel: you can change the position of the slices with the three sliders. == Surfaces visualization == There is only one way to display the surfaces: in 3D figures. To display a surface you can either double-click on it or right-click > Display. * The mouse and keyboard operations are the same described for the 3D MRI display also apply here. * If you display two surfaces from the same subject, they will be displayed on the same figure. * Surfaces tab: more options are available<<BR>><<BR>> {{attachment:panelSurfaces.gif}} * Open the ''Cortex ''surface and try all the buttons and sliders. Then do the same with the ''Scalp ''surface (''Head''). All the controls should have some effect on the display, except the sliders in the "''Data options''" panel. They are useful only when some additional data is mapped on the surfaces. * Multiple surfaces: If you have more than one surface in a figure, you need to select the surface you want to edit before changing its properties. The list of the available surfaces is displayed on top of the ''Surfaces ''tab. * You can also quickly add or remove surfaces in any 3D figure with the top-right buttons of the ''Surfaces ''tab. == Coordinates tab == * Close all the figures. Open the cortex surface again. * Right-click on the 3D figure, select "Get coordinates". A new window appears. * Click anywhere on the cortex surface: a big yellow cross appears, and the coordinates of the point are displayed in all the available coordinates systems (see page: CoordinateSystems) * MRI: MR volume, in millimeters (multiplied with the Voxsize field in the MRI file). * SCS: Subject Coordinates System, in millimeters * MNI: Montreal Neurological Institute standard coordinates * You can click on "View / MRI" to see where this point is located in the MRI, using the MRI Viewer. <<BR>><<BR>> {{attachment:panelCoord.gif}} {{attachment:panelCoordPt.gif}} == Create a subject == If you explored well, you should have found the "''New subject''" menu in the protocol's popup menu. ||<tablewidth="200px"#ffffff style="vertical-align:top; "> {{attachment:menuNewSubject.gif}} || {{attachment:createNewSubject.gif}} || * Usually, the only thing you need to do here is to edit the name of the subject. * But you can also override here the protocol's defaults regarding the sharing of anatomy and channel file between subjects and conditions. For the example, leave selected the "''Yes, use default anatomy''" option. Then click on ''Save''. * You should see a the new subject in both ''Anatomy ''and ''Functional data'' views. * In the Anatomy view, the subject node only contains a link called (Default anatomy). It is here just to remind that the subject is using the default ''MNI Colin27'' anatomy.<<BR>><<BR>> {{attachment:subjectDefaultAnat.gif}} == Change the default anatomy == When you create a new protocol, the program makes a copy of the Colin27 anatomy and sets it as the default for the protocol. It means that you will be able to use the Colin27 brain as a substitute for the subjects without an individual MRI, or as the common brain for group analysis. Other sets of MRI+surfaces are available to replace the Colin27 anatomy. Right-click on ''(Default anatomy)'' > Use template. If a package is not currently available on your system, it will be downloaded from the Brainstorm website and saved in $HOME/.brainstorm/templates. The available options are: * '''Colin27''': Average of 27 scans of the same head, processed with FreeSurfer 5.3: [[http://www.bic.mni.mcgill.ca/ServicesAtlases/Colin27|more information]] * '''Colin27_2012''': Previous version of the default anatomy distributed with Brainstorm * '''ICBM152''': Non-linear average of 152 subjects, processed with FreeSurfer 5.3: [[http://www.bic.mni.mcgill.ca/ServicesAtlases/ICBM152NLin2009|more information]] * '''FSAverage''': Average of 40 subjects using a spherical averaging described in [[http://nmr.mgh.harvard.edu/~fischl/reprints/morphing_human_brain_mapping_reprint.pdf|(Fischl et al. 1999)]].<<BR>>It is the default FreeSurfer brain: please [[https://surfer.nmr.mgh.harvard.edu/registration.html|register here]] if you are using it. * '''Infant7w''': 7-week infant brain with the antomical atlas presented in [[http://www.sciencedirect.com/science/article/pii/S105381191400411X|(Kabdebon et al. 2014)]]. They all include the following information: * T1 MRI volume * Cortex surface: high-resolution (~300.000 vertices) and low-resolution (15.000 vertices) * Head surface: based on the head used for FSAverage in the MNE software * FreeSurfer spherical registration of each hemisphere, with which we can co-register the individual brains processed with FreeSurfer with the selected default anatomy * FreeSurfer surface-based atlases: Desikan-Killiany, Destrieux, Brodman, Mindboggle<<BR>>(plus Yeo2011 and PALS for FSAverage only) The atlases will be discussed in the following tutorials. For more information on the interactions between FreeSurfer and Brainstorm: [[Tutorials/LabelFreeSurfer|read this tutorial]]. {{attachment:changeDefault.gif|changeDefault1.gif}} If you click on any of the download options, it downloads it into your $HOME/.brainstorm/templates folder: {{attachment:changeDefault2.gif}} Then the list of files in the (default anatomy) folder is replaced with the new template. {{attachment:changeDefault3.gif}} == Modify the default MRI fiducials == The fiducial points (Nasion, LPA, RPA) used in your recordings might not be the same as the ones used in the anatomy templates in Brainstorm (Colin27, ICBM152, FSAverage). By default, the LPA/RPA points are defined at the junction between the tragus and the helix, as represented with the red dot in the [[http://neuroimage.usc.edu/brainstorm/CoordinateSystems|Coordinates systems page]]. If you want to use an anatomy template but you are using a different convention when digitizing the position of those points, you have to modify the default positions of the template with the MRI Viewer. * Go to the anatomy view * In (default anatomy), right-click on the MRI > Edit MRI * Modify the position of the fiducial points to match your own convention * Click on [Save], it will update the surfaces to match the new coordinate system == Next == Now you know how to create a protocol with a default anatomy, and visualize MR volumes and surfaces. Next step: how to [[Tutorials/TutImportAnatomy|import an individual anatomy]] for a subject. <<EmbedContent("http://neuroimage.usc.edu/bst/get_prevnext.php?prev=Tutorials&next=Tutorials/TutFirstSteps")>> |
<<EmbedContent("http://neuroimage.usc.edu/bst/get_prevnext.php?prev=Tutorials&next=Tutorials/ImportAnatomy")>> |
Tutorial 1: Create a new protocol
Authors: Francois Tadel, Elizabeth Bock, Sylvain Baillet
Contents
How to read these tutorials
The goal of these introduction tutorials is to guide you through most of the features of the software. All the pages use the same example dataset. The results of one section are most of the time used in the following section, so read these pages in the correct order.
Some pages may contain too many details for your level of interest or competence. The sections marked as [Advanced] are not required for you to follow the tutorials until the end. You can skip them the first time you go through the documentation. You will be able to get back to the theory later if you need.
Please follow first these tutorials with the data we provide. This way you will be able to focus on learning how to use the software. It is better to start with some data that is easy to analyze. After going through all the tutorials, you should be comfortable enough with the software to start analyzing your own recordings.
If you are interested only in EEG or intra-cranial recordings, don't think that an MEG-based tutorial is not adapted for you. Most of the practical aspects of the data manipulation is very similar in EEG and MEG. Start by reading these introduction tutorials, then go through the tutorial "EEG and Epilepsy" to get some more details about the processing steps that are specific for EEG.
Presentation of the experiment
All the introduction tutorials are based on a simple auditory oddball experiment:
- One subject, two acquisition runs of 6 minutes each
- Subject stimulated binaurally with intra-aural earphones
- Each run contains 200 regular beeps and 40 easy deviant beeps
- Recordings with a CTF MEG system with 275 axial gradiometers
Anatomy of the subject: 1.5T MRI, processed with FreeSurfer 5.3
- More details will be given about this dataset along the process.
Full dataset description available on this page: Introduction dataset
Brainstorm folders
Brainstorm needs different directories to work properly. If you put everything in the same folder, you would run into many problems. Try to understand this organization before creating a new database.
1. Program directory: "brainstorm3"
- Contains all the program files: Matlab scripts, compiled binaries, templates, etc.
- There is no user data in this folder.
- You can delete it and replace it with a newer version at anytime, your data will be safe.
- Recommended location:
Windows: My documents\brainstorm3
Linux: /home/username/brainstorm3
MacOS: Documents/brainstorm3
2. Database directory: "brainstorm_db"
- Created by user.
- Contains all the Brainstorm database files.
- Managed automatically by the application: do not move, delete or add files by yourself.
- Recommended location:
Windows: My documents\brainstorm_db
Linux: /home/username/brainstorm_db
MacOS: Documents/brainstorm_db
3. User directory: ".brainstorm"
Created automatically at Brainstorm startup. Typical location:
Windows: C:\Documents and Settings\username\.brainstorm
Linux: /home/username/.brainstorm
MacOS: /Users/username/.brainstorm
- Contains:
brainstorm.mat: Brainstorm user preferences.
tmp/: Temporary folder, cleaned every time Brainstorm is started.
You may have to change the location of the temporary folder if you have a limited amount of storage or a limited quota in your home folder.process/: Personal plugins folder.
reports/: Process reports.
openmeeg/: OpenMEEG binaries (downloaded automatically when needed)
templates/: Additional anatomy templates, saved as .zip files.
mex/: Some mex files that have to be recompiled.
4. Original data files:
- Recordings you acquired and you want to process with Brainstorm.
Put them wherever you want but not in any of the previous folders.
Starting Brainstorm for the first time
If you haven't read the installation instructions, do it now: Installation.
Start Brainstorm from Matlab or with the compiled executable.
BST> Starting Brainstorm: BST> ================================= BST> Version: 28-Jan-2015 BST> Checking internet connectivity... ok BST> Compiling main interface files... BST> Emptying temporary directory... BST> Deleting old process reports... BST> Loading configuration file... BST> Initializing user interface... BST> Starting OpenGL engine... hardware BST> Reading plugins folder... BST> Loading current protocol... BST> =================================
- Read and accept the license file.
- Select your Brainstorm database directory (brainstorm_db).
If you do something wrong and don't know how to go back, you can always re-initialize Brainstorm by typing "brainstorm reset" in the Matlab command window, or by clicking on [Reset] in the software preferences (menu File > Edit preferences).
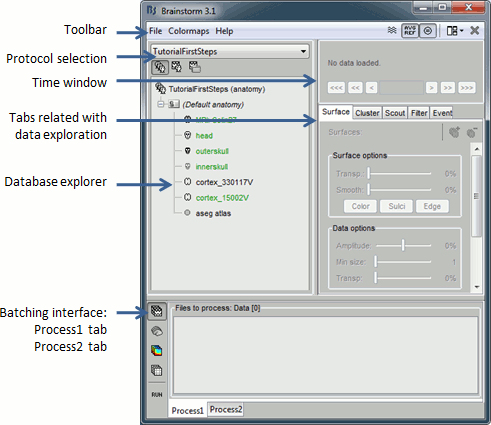
Main interface window
The Brainstorm window described below is designed to remain on one side of the screen. All the space of the desktop that is not covered by this window will be used for opening other figures.
Do not try to maximize this window, or the automatic management of the data figures might not work correctly. Keep it on one side of your screen, just large enough so you can read the file names in the database explorer.
Database structure
Brainstorm allows you to organize your recordings and analysis with three levels of definition:
Protocol
- Group of datasets that have to be processed or displayed together.
- A protocol can include one or several subjects.
Some people would prefer to call this experiment or study.
- You can only open one protocol at a time.
- Your Brainstorm database is a collection of protocols.
Subject
- A person who participated in a given protocol.
- A subject contains two categories of information: anatomy and functional data.
Anatomy: Includes at least an MRI volume and some surfaces extracted from the MRI.
Functional data: Everything that is related with the MEG/EEG acquisition.
- For each subject, it is possible to use either the actual MRI of the person or one of the anatomy templates available in Brainstorm.
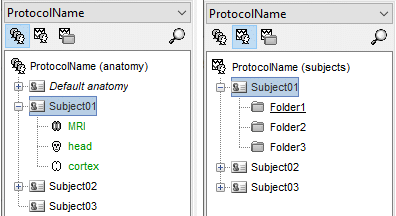
Sub-folders
- For each subject, the functional files can be organized in different sub-folders.
- These folders can represent different recordings sessions (aka acquisition runs) or different experimental conditions.
The current structure of the database does not allow more than one level of sub-folders for each subject. It is not possible to organize the files by session AND by condition.
Database files
- The database folder "brainstorm_db" is managed completely from the graphic user interface (GUI).
- All the files in the database have to be imported through the GUI. Do not try to copy files by yourself in the brainstorm_db folder, it won't work.
- Everything in this folder is stored in Matlab .mat format, with the following architecture:
Anatomy data: brainstorm_db/protocol_name/anat/subject_name
Functional data: brainstorm_db/protocol_name/data/subject_name/subfolder/
- Most of the files you see in the database explorer in Brainstorm correspond to files on the hard drive, but there is no one-to-one correspondence. There is extra information stored in each directory, to save properties, comments, default data, links between different items, etc. This is one of the reasons for which you should not try to manipulate directly the files in the Brainstorm database directory.
- The structure of the database is saved in the user preferences, so when you start the program or change protocol, there is no need to read again all the files on the hard drive.
If Brainstorm or Matlab crashes before the database structure is correctly saved, the files that are displayed in the Brainstorm database explorer may differ from what is actually on the disk. When this happens, you can force Brainstorm to rebuild the structure from the files on the hard drive: right-click on a folder > Reload.
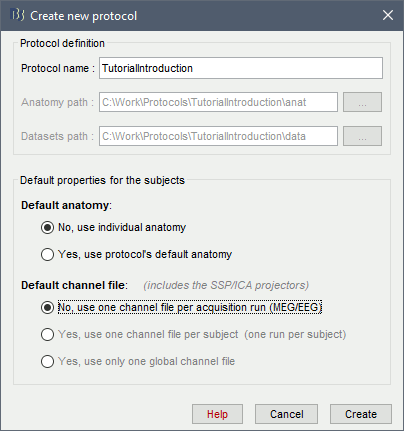
Create your first protocol
Menu File > New protocol.
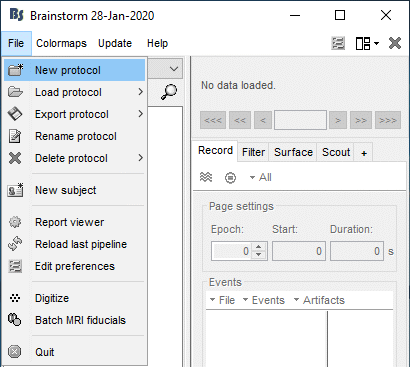
Edit the protocol name and enter: "TutorialIntroduction".
It will automatically update the anatomy and datasets paths.- Default properties for the subjects: These are the default settings that are used when creating new subjects. It is then possible to override these settings for each subject individually.
Default anatomy: (MRI and surfaces)
No, use individual anatomy:
Select when you have individual MRI scans for all the participants of your study.Yes, use default anatomy:
Select when you do not have individual scans for the participants, and you would like to use one of the anatomy templates available in Brainstorm.
Default channel file: (Sensors names and positions)
No, use one channel file per condition/run:
Select this if you may have different head positions for one subject. This is usually not the case in EEG, where the electrodes stay in place for all the experiment. In MEG, this is a common setting: one recording session is split in multiple acquisition runs, and the position of the subject's head in the MEG might be different between two runs.Yes, use one channel file per subject:
This is a common setting for EEG. The electrodes are in the same position for all the files recorded on one subject.Yes, use only one global channel file:
This is never a recommended setting. It could be used in the case of an EEG study where you use only standard EEG positions on a standard anatomy, but only if you are not doing any advanced source reconstruction. If you share the position of the electrodes between all the subjects, it will also share the source models, which are dependent on the quality of the recordings for each subject. This is complicated to understand at this level, it will make more sense later in the tutorials.
In the context of this study, we are going to use the following settings:
No, use individual anatomy: Because we have access to a T1 MRI scan of the subject.
No, use one channel file per condition/run: The typical MEG setup.
Click on [Create].
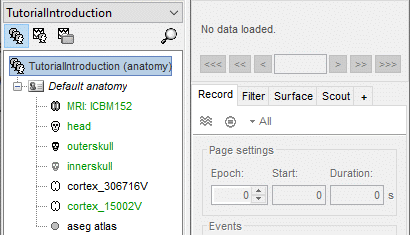
Protocol exploration
The protocol is created and you can now see it in the database explorer. It is represented by the top-most node in the tree.
- You can switch between anatomy and functional data with the three buttons just above the database explorer. Read the tooltips of the buttons to see which one does what.
In the Anatomy view, there is a Default anatomy node. It contains the Colin27 anatomy, distributed by the Montreal Neurological Institute (MNI), which is one of the template anatomy folders that are distributed with Brainstorm.
The Default anatomy node contains the MRI and the surfaces that are used for the subjects without an individual anatomy, or for registering multiple subjects to the same anatomy for group analysis.
There are no subjects in the database yet, so the Functional data views are empty.
Everything you can do with an object in the database explorer (anatomy files, subjects, protocol) is accessible by right-clicking on it.
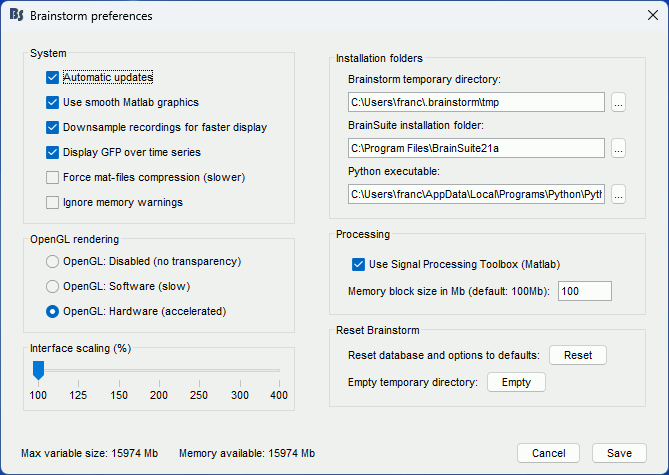
Changing the temporary folder
If the amount of storage space you have for your user folder is limited (less than 10Gb), you may have to change the temporary folder used by Brainstorm. If you work on a centralized network where all the computers are sharing the same resources, the system admins may impose limited disk quotas to all users and encourage them to use local hard drives instead of the limited and shared user folder.
The default temporary folder of Brainstorm is located in your user folder ($HOME/.brainstorm/tmp/), so when you import recordings or calculate large models, it may quickly fill up your limited quota and at some point block your user account. To prevent this, select the menu File > Edit preferences and set the temporary directory to a folder that is local to your computer, in which you won't suffer from any storage limitation.
Summary
- Different folders for:
the program (brainstorm3),
the database (brainstorm_db),
your original recordings.
- Never modify the contents of the database folder by yourself
- Do not put the original recordings in any of the Brainstorm folders, import them with the interface
- Do not try to maximize the Brainstorm window: keep it small on one side of your screen.