|
Size: 7728
Comment:
|
Size: 7157
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Decoding conditions = ''Authors: Seyed-Mahdi Khaligh-Razavi, Francois Tadel, Dimitrios Pantazis'' |
= Machine learning: Decoding / MVPA = ''Authors: Dimitrios Pantazis''', '''Seyed-Mahdi Khaligh-Razavi, Francois Tadel, '' |
| Line 4: | Line 4: |
| This tutorial illustrates how to use the functions developed at Aude Oliva's lab (MIT, CSAIL), and McGovern's MEG lab (Dimitrios Pantazis's lab) to run support vector machine (SVM) and linear discriminant analysis (LDA) classification on your MEG data across time. | This tutorial illustrates how to run MEG decoding using support vector machines (SVM). |
| Line 8: | Line 8: |
| <<Include(DatasetDecoding, , from="\<\<HTML\(\<!-- START-PAGE --\>\)\>\>", to="\<\<HTML\(\<!-- STOP-SHORT --\>\)\>\>")>> | == License == To reference this dataset in your publications, please cite Cichy et al. (2014). |
| Line 13: | Line 14: |
| * Decoding > Classification with cross-validation (process_decoding_crossval.m) * Decoding > Classification with permutation (process_decoding_permutation.m) |
* Decoding > Classification with SVD * Decoding > Classification with max-correlation classifier |
| Line 16: | Line 17: |
| These two processes work in a similar way: | These two processes work in a similar way, but they use a different classifier, so only SVD is demonstrated here. |
| Line 18: | Line 19: |
| * '''Input''': the input is the channel data from two conditions (e.g. condA and condB) across time. Number of samples per condition should be the same for both condA and condB. Each of them should at least contain two samples. * '''Output''': the output is a decoding curve across time, showing your decoding accuracy (decoding condA vs. condB) at time point 't'. * '''Classifier''': Two methods are offered for the classification of MEG recordings across time: Support vector machine (SVM) and Linear discriminant analysis (LDA). |
* '''Input''': the input is the channel data from two conditions (e.g. condA and condB) across time. Number of samples per condition do not have to be the same for both condA and condB, but each of them should have enough samples to create k-folds (see parameter below). * '''Output''': the output is a decoding time course, or a temporal generalization matrix (train time x test time). * '''Classifier''': Two methods are offered for the classification of MEG recordings across time: support vector machine (SVM) and max-correlation classifier. |
| Line 22: | Line 23: |
| In the context of this tutorial, we have two condition types: faces, and scenes. We want to decode faces vs. scenes using 306 MEG channels. In the data, the faces are named as condition ‘201’; and the scenes are named as condition ‘203’. {{attachment:decoding_protocol.gif}} |
In the context of this tutorial, we have two condition types: faces, and objects. We want to decode faces vs. objects using 306 MEG channels. |
| Line 27: | Line 26: |
| * Go to the [[http://neuroimage.usc.edu/bst/download.php|Download]] page of this website, and download the file: '''sample_decoding.zip ''' * Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder). This is really important that you always keep your original data files in a separate folder: the program folder can be deleted when updating the software, and the contents of the database folder is supposed to be manipulated only by the program itself. |
* From the [[http://neuroimage.usc.edu/bst/download.php|Download]] page of this website, download the file: '''subj04NN_sess01-0_tsss.fif''' |
| Line 33: | Line 31: |
| * "'''No, use one channel file per condition'''". <<BR>><<BR>> {{attachment:decoding_protocol.gif||width="344",height="370"}} | * "'''No, use one channel file per condition'''". <<BR>><<BR>> {{attachment:decoding_protocol.gif||height="370",width="344"}} |
| Line 38: | Line 36: |
| * Right click on the subject node (Subject01) > '''Review raw file'''''.'' <<BR>>Select the file format: "'''MEG/EEG: Neuromag FIFF (*.fif)'''"<<BR>>Select the file: sample_decoding/'''mem6-0_tsss_mc.fif''' <<BR>><<BR>> {{attachment:decoding_link.gif||width="547",height="169"}} * Select "Event channels" to read the triggers from the stimulus channel. <<BR>><<BR>> {{attachment:decoding_events.gif||width="289",height="162"}} |
* Right click on the subject node (Subject01) > '''Review raw file'''''.'' <<BR>>Select the file format: "'''MEG/EEG: Neuromag FIFF (*.fif)'''"<<BR>>Select the file: sample_decoding/'''mem6-0_tsss_mc.fif''' <<BR>><<BR>> {{attachment:decoding_link.gif||height="169",width="547"}} * Select "Event channels" to read the triggers from the stimulus channel. <<BR>><<BR>> {{attachment:decoding_events.gif||height="162",width="289"}} |
| Line 41: | Line 39: |
| * Right-click on the "Link to raw file" > '''Import in database'''. <<BR>><<BR>> {{attachment:decoding_import.gif||width="571",height="317"}} | * Right-click on the "Link to raw file" > '''Import in database'''. <<BR>><<BR>> {{attachment:decoding_import.gif||height="317",width="571"}} |
| Line 53: | Line 51: |
| * You can select more than 40 or less. The important thing is that both ‘A’ and ‘B’ should have the same number of files. <<BR>><<BR>> {{attachment:decoding_selectfiles.gif||width="398",height="369"}} | * You can select more than 40 or less. The important thing is that both ‘A’ and ‘B’ should have the same number of files. <<BR>><<BR>> {{attachment:decoding_selectfiles.gif||height="369",width="398"}} |
| Line 58: | Line 56: |
| * Select process "'''Decoding > Classification with cross-validation'''": <<BR>><<BR>> {{attachment:cv_process.gif||width="346",height="388"}} | * Select process "'''Decoding > Classification with cross-validation'''": <<BR>><<BR>> {{attachment:cv_process.gif||height="388",width="346"}} |
| Line 63: | Line 61: |
| * The process will take some time. The results are then saved in a file in the new decoding folder. <<BR>><<BR>> {{attachment:cv_file.gif||width="268",height="167"}} * If you double click on it you will see a decoding curve across time. <<BR>><<BR>> {{attachment:cv_plot.gif||width="355",height="172"}} |
* The process will take some time. The results are then saved in a file in the new decoding folder. <<BR>><<BR>> {{attachment:cv_file.gif||height="167",width="268"}} * If you double click on it you will see a decoding curve across time. <<BR>><<BR>> {{attachment:cv_plot.gif||height="172",width="355"}} |
| Line 69: | Line 67: |
| * Select process "'''Decoding > Classification with cross-validation'''". Set options as below: <<BR>><<BR>> {{attachment:perm_process.gif||width="311",height="366"}} | * Select process "'''Decoding > Classification with cross-validation'''". Set options as below: <<BR>><<BR>> {{attachment:perm_process.gif||height="366",width="311"}} |
| Line 71: | Line 69: |
| * The results are saved in a file in the new decoding folder. <<BR>><<BR>> {{attachment:perm_file.gif||width="284",height="142"}} * Right-click > Display as time series (or double click). <<BR>><<BR>> {{attachment:perm_plot.gif||width="346",height="169"}} |
* The results are saved in a file in the new decoding folder. <<BR>><<BR>> {{attachment:perm_file.gif||height="142",width="284"}} * Right-click > Display as time series (or double click). <<BR>><<BR>> {{attachment:perm_plot.gif||height="169",width="346"}} |
| Line 81: | Line 79: |
| == Additional documentation == * Forum: Decoding in source space: http://neuroimage.usc.edu/forums/showthread.php?2719 |
Machine learning: Decoding / MVPA
Authors: Dimitrios Pantazis, Seyed-Mahdi Khaligh-Razavi, Francois Tadel,
This tutorial illustrates how to run MEG decoding using support vector machines (SVM).
Contents
License
To reference this dataset in your publications, please cite Cichy et al. (2014).
Description of the decoding functions
Two decoding processes are available in Brainstorm:
Decoding > Classification with SVD
Decoding > Classification with max-correlation classifier
These two processes work in a similar way, but they use a different classifier, so only SVD is demonstrated here.
Input: the input is the channel data from two conditions (e.g. condA and condB) across time. Number of samples per condition do not have to be the same for both condA and condB, but each of them should have enough samples to create k-folds (see parameter below).
Output: the output is a decoding time course, or a temporal generalization matrix (train time x test time).
Classifier: Two methods are offered for the classification of MEG recordings across time: support vector machine (SVM) and max-correlation classifier.
In the context of this tutorial, we have two condition types: faces, and objects. We want to decode faces vs. objects using 306 MEG channels.
Download and installation
From the Download page of this website, download the file: subj04NN_sess01-0_tsss.fif
- Start Brainstorm (Matlab scripts or stand-alone version).
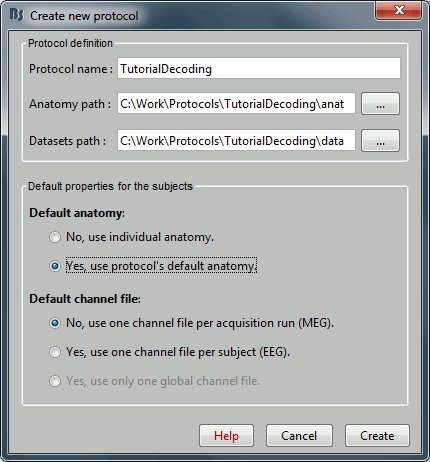
Select the menu File > Create new protocol. Name it "TutorialDecoding" and select the options:
"Yes, use protocol's default anatomy",
"No, use one channel file per condition".
Import the recordings
- Go to the "functional data" view (sorted by subjects).
Right-click on the TutorialDecoding folder > New subject > Subject01
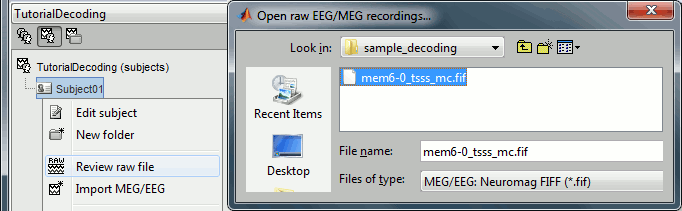
Leave the default options you defined for the protocol.Right click on the subject node (Subject01) > Review raw file.
Select the file format: "MEG/EEG: Neuromag FIFF (*.fif)"
Select the file: sample_decoding/mem6-0_tsss_mc.fif
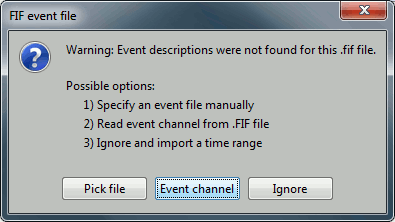
Select "Event channels" to read the triggers from the stimulus channel.
- We will not pay much attention to MEG/MRI registration because we are not going to compute any source models, the decoding is done on the sensor data.
Right-click on the "Link to raw file" > Import in database.
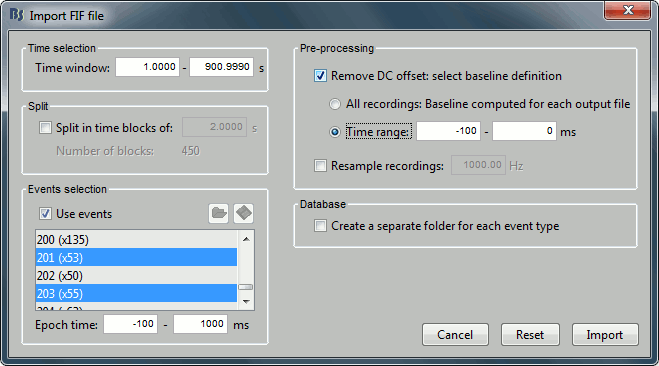
- Select only two events: 201 (faces) and 203(scenes)
- Epoch time: [-100, 1000] ms
- Remove DC offset: Time range: [-100, 0] ms
- Do not create separate folders for each event type
You will get a message saying "some epochs are shorter than the others". Answer yes.
Select files
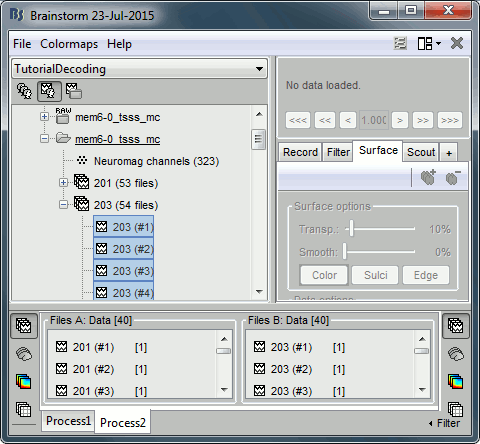
Select the Process2 tab at the bottom of the Brainstorm window.
Drag and drop 40 files from group 201 to the left (Files A).
Drag and drop 40 files from group 203 to the right (Files B).
You can select more than 40 or less. The important thing is that both ‘A’ and ‘B’ should have the same number of files.
Cross-validation
Cross-validation is a model validation technique for assessing how the results of our decoding analysis will generalize to an independent data set.
Select process "Decoding > Classification with cross-validation":
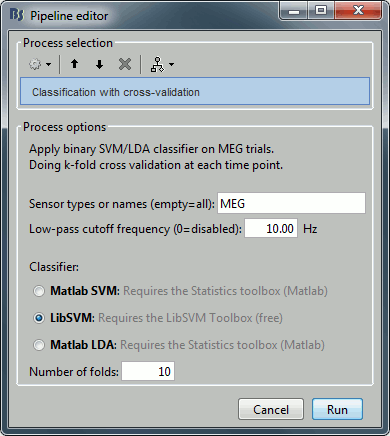
Low-pass cutoff frequency: If set, it will apply a low-pass filter to all the input recordings.
Matlab SVM/LDA: Require Matlab's Statistics and Machine Learning Toolbox.
These methods do a k-fold stratified cross-validation for you, meaning that each fold will contain the same proportions of the two types of class labels (option "Number of folds").LibSVM: Requires the LibSVM toolbox (download and add to your path).
The LibSVM cross-validation may be faster but will not be stratified.
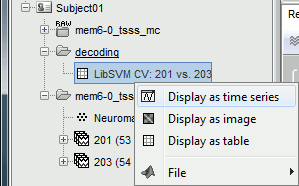
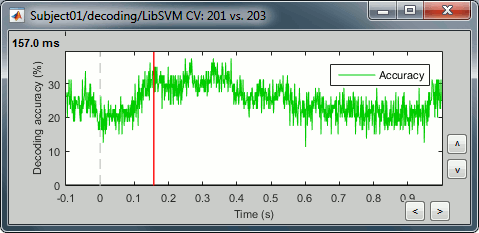
The process will take some time. The results are then saved in a file in the new decoding folder.
If you double click on it you will see a decoding curve across time.
Permutation
This is an iterative procedure. The training and test data for the SVM/LDA classifier are selected in each iteration by randomly permuting the samples and grouping them into bins of size n (you can select the trial bin sizes). In each iteration two samples (one from each condition) are left out for test. The rest of the data are used to train the classifier with.
Select process "Decoding > Classification with cross-validation". Set options as below:
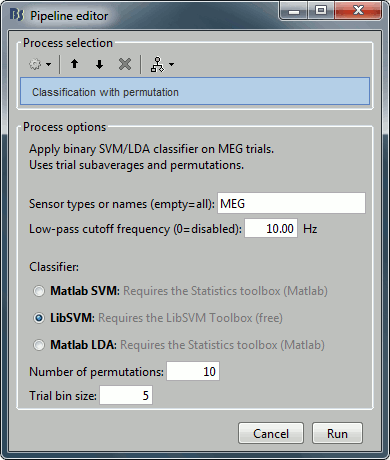
Trial bin size: If greater than 1, the training data will be randomly grouped into bins of the size you determine here. The samples within each bin are then averaged (we refer to this as sub-averaging); the classifier is then trained using the averaged samples. For example, if you have 40 faces and 40 scenes, and you set the trial bin size to 5; then for each condition you will have 8 bins each containing 5 samples. Seven bins from each condition will be used for training, and the two left-out bins (one face bin, one scene bin) will be used for testing the classifier performance.
The results are saved in a file in the new decoding folder.
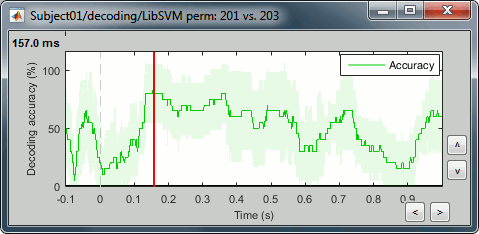
Right-click > Display as time series (or double click).
Acknowledgment
This work was supported by the McGovern Institute Neurotechnology Program to PIs: Aude Oliva and Dimitrios Pantazis. http://mcgovern.mit.edu/technology/neurotechnology-program
References
Khaligh-Razavi SM, Bainbridge W, Pantazis D, Oliva A (2016)
From what we perceive to what we remember: Characterizing representational dynamics of visual memorability. bioRxiv, 049700.Cichy RM, Pantazis D, Oliva A (2014)
Resolving human object recognition in space and time, Nature Neuroscience, 17:455–462.
Additional documentation
Forum: Decoding in source space: http://neuroimage.usc.edu/forums/showthread.php?2719
