|
Size: 4391
Comment:
|
Size: 6679
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 24: | Line 24: |
| * Create one subject. * Right-click on the subject node > Import anatomy folder > sample_epilepsy/anatomy (format=FreeSurfer) * For now the MRI is processed with BrainVISA, but we would like to have it fully processed with !FreeSurfer. * If no anatomy available: [[Tutorials/TutWarping|Warping tutorial]] |
* Right-click on the !TutorialEpilepsy folder > New subject > '''sepi01''' * Leave the default options you set for the protocol * Right-click on the subject node > Import anatomy folder: * Set the file format: "!FreeSurfer folder" * Select the folder: '''sample_epilepsy/anatomy''' * Number of vertices of the cortex surface: 15000 (default value) * Set the 6 required fiducial points (indicated in MRI coordinates): * NAS: x=135, y=222, z=75 * LPA: x=57, y=118, z=68 * RPA: x=204, y=119, z=76 * AC: x=131, y=145, z=110 * PC: x=130, y=119, z=111 * IH: x=128, y=134, z=170 (anywhere on the midsagittal plane) {{attachment:anatomy.gif||height="257",width="442"}} ==== Without the individual MRI ==== If you do not have access to an individual MR scan of the subject (or if its quality is too low to be processed with !FreeSurfer), but if you have digitized the head shape of the subject using a tracking system, you have an alternative: deform one of the Brainstorm templates (Colin27 or ICBM152) to match the shape of the subject's head.<<BR>>For more information, read the following tutorial: [[Tutorials/TutWarping|Warping default anatomy]] |
| Line 31: | Line 44: |
| * Right-click on the subject folder > Review raw file > sample_epilepsy/data/tutorial_EEG.bin (format = "EEG: Deltamet Neurofile-Coherence (*.bin)") * Right-click on the subject folder > Import channel file > sample_epilepsy/data/XensorTest.elc (format = "EEG: ANT Xensor (*.elc)")<<BR>=> Contains the default electrodes positions from the ASA software (ANT). * Change the sensor type of the following electrodes to '''MISC''': SP1, SP2, RS, PHO, DELR, DELL, QR, QL |
* Right-click on the subject folder > Review raw file: * Select the file format: "EEG: Deltamet Neurofile-Coherence (*.bin)" * Select the file: '''sample_epilepsy/data/tutorial_EEG.bin''' * The new file "Link to raw file" lets you access directly the contents of the EEG recordings * The channel file "Deltamed channels" in the (Common files) folder contains the name of the channels, but not their positions. We need to overwrite this file and import manually the positions of the electrodes (either a standard cap or accurate positions digitized with a Polhemus device). * Right-click on the subject folder > Import channel file: * Select the file format: "EEG: ANT Xensor (*.elc)" * Select the file: '''sample_epilepsy/data/tutorial.elc''' * Confirm that you want to overwrite the exising channel file. * This file contains the default electrodes positions from the ASA software (ANT) * The recordings contain signals coming from different types of electrodes: * 29 EEG electrodes * EOG1, EOG2: Electrooculograms * EMG, ECG: Electromyogram and electrocardiogram * SP1, SP2: Sphenoidal electrodes * RS: ? * PHO: ? * DELR, DELL: ? * QR, QL: ? * The file format for the electrodes positions does not describe the type of the channels correctly, therefore all the signals saved in the files are classified as EEG. We need to redefine this manually to get correct groups of sensors, we want to have only real EEG electrodes in the "EEG" category and put everything that we are not going to use a "MISC" category. * Right-click on the channel file > Edit channel file: * Note that the EOG, EMG and ECG channels already have a different type * Select all the other non-EEG channels: SP1, SP2, RS, PHO, DELR, DELL, QR, QL * Right-click in the window > Set channel type: type '''MISC''' {{attachment:channel_type.gif}} |
EEG and epilepsy
This tutorial introduces some concepts that are specific to the management of EEG recordings in the Brainstorm environment. It also describes a standard pipeline for analyzing epilepsy recordings. It is based on clinical case from the Epilepsy Centre, at the University Hospital Freiburg, Germany. The EEG data was recorded at 1024Hz, using a Neurofile NT digital video-EEG system with 128 channels. The anonymized dataset can be downloaded directly from the Brainstorm download page.
The case from this tutorial is also published in this article: Dümpelmann M, Ball T, Schulze-Bonhage A
LORETA allows reliable distributed source reconstruction based on subdural strip and grid recordings, Hum Brain Mapp. 2012
License
This tutorial dataset (EEG and MRI data) remains proprietary of the Epilepsy Centre, University Hospital Freiburg, Germany. Its use and transfer outside the Brainstorm tutorial, e.g. for research purposes, is prohibited without written consent from the Epilepsy Centre in Freiburg. For questions please contact A. Schulze-Bonhage, MD, PhD: andreas.schulze-bonhage@uniklinik-freiburg.de
Presentation of the clinical case
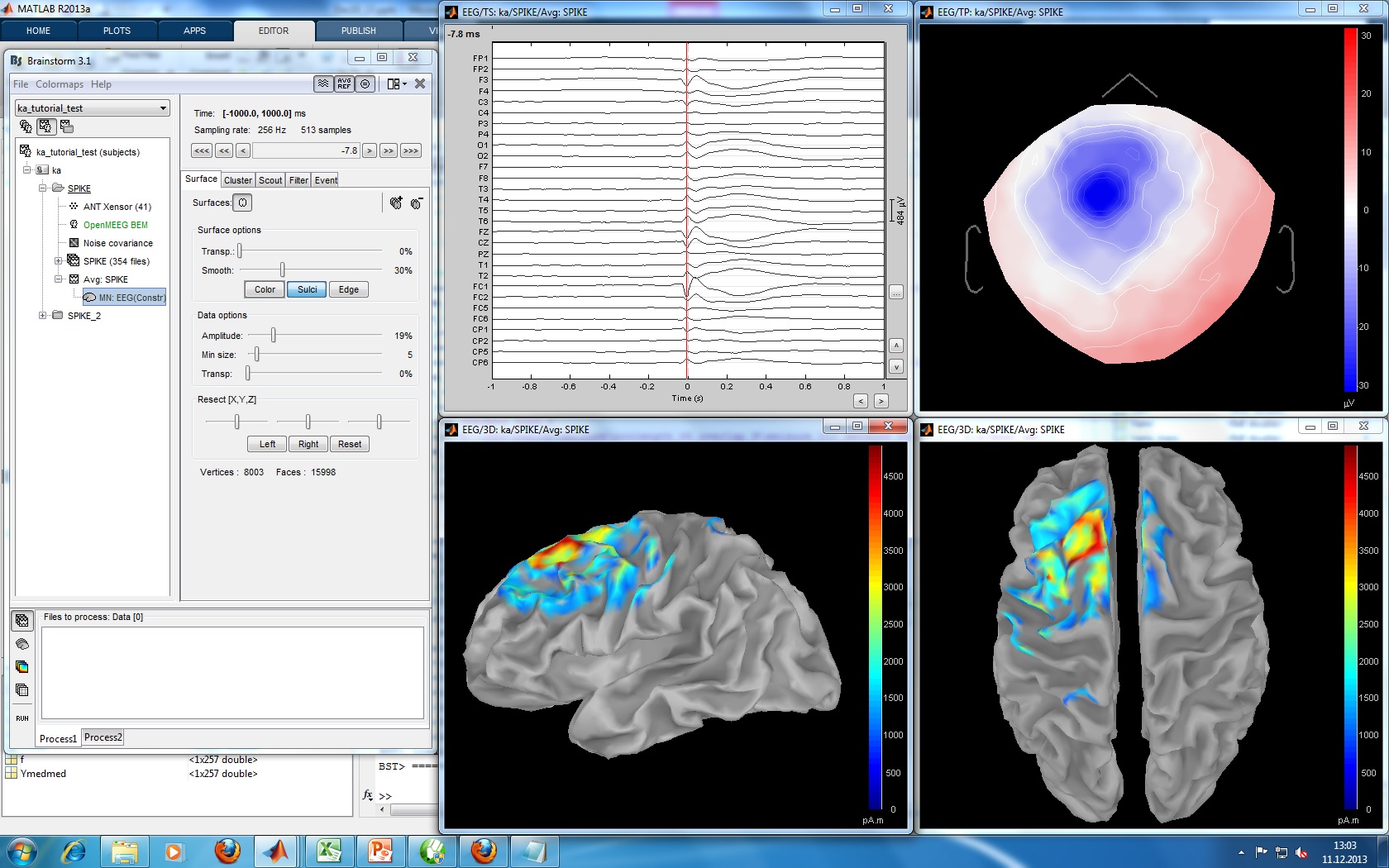
The EEG data was recorded at 1024Hz, using a Neurofile NT digital video-EEG system with 128 channels and a 16-bit A/D converter. The signal was filtered in the recording system with a high-pass filter with a time constant of 1 second and a low-pass filter with a cutoff frequency of 344 Hz. The spikes were visually identified and averaged with the ASA package. The spike average showed prominent peaks in the grid contacts G_A2-4, G_B2-5, G_C1-3.
Type of epilepsy, supposed location, clinical conclusions, etc.
Download and installation
- Requirements: You have already followed all the introduction tutorials and you have a working copy of Brainstorm installed on your computer.
Go to the Download page of this website, and dowload the file: sample_epilepsy.zip
Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder)
- Start Brainstorm (Matlab scripts or stand-alone version)
Select the menu File > Create new protocol. Name it "TutorialEpilepsy" and select the options:
"No, use individual anatomy",
"Yes, use one channel file per subject".
Import the anatomy
Right-click on the TutorialEpilepsy folder > New subject > sepi01
- Leave the default options you set for the protocol
Right-click on the subject node > Import anatomy folder:
Set the file format: "FreeSurfer folder"
Select the folder: sample_epilepsy/anatomy
- Number of vertices of the cortex surface: 15000 (default value)
- Set the 6 required fiducial points (indicated in MRI coordinates):
- NAS: x=135, y=222, z=75
- LPA: x=57, y=118, z=68
- RPA: x=204, y=119, z=76
- AC: x=131, y=145, z=110
- PC: x=130, y=119, z=111
- IH: x=128, y=134, z=170 (anywhere on the midsagittal plane)
Without the individual MRI
If you do not have access to an individual MR scan of the subject (or if its quality is too low to be processed with FreeSurfer), but if you have digitized the head shape of the subject using a tracking system, you have an alternative: deform one of the Brainstorm templates (Colin27 or ICBM152) to match the shape of the subject's head.
For more information, read the following tutorial: Warping default anatomy
Access the recordings
- Switch to the "functional data" view.
Right-click on the subject folder > Review raw file:
- Select the file format: "EEG: Deltamet Neurofile-Coherence (*.bin)"
Select the file: sample_epilepsy/data/tutorial_EEG.bin
- The new file "Link to raw file" lets you access directly the contents of the EEG recordings
- The channel file "Deltamed channels" in the (Common files) folder contains the name of the channels, but not their positions. We need to overwrite this file and import manually the positions of the electrodes (either a standard cap or accurate positions digitized with a Polhemus device).
Right-click on the subject folder > Import channel file:
- Select the file format: "EEG: ANT Xensor (*.elc)"
Select the file: sample_epilepsy/data/tutorial.elc
- Confirm that you want to overwrite the exising channel file.
- This file contains the default electrodes positions from the ASA software (ANT)
- The recordings contain signals coming from different types of electrodes:
- 29 EEG electrodes
- EOG1, EOG2: Electrooculograms
- EMG, ECG: Electromyogram and electrocardiogram
- SP1, SP2: Sphenoidal electrodes
- RS: ?
- PHO: ?
- DELR, DELL: ?
- QR, QL: ?
- The file format for the electrodes positions does not describe the type of the channels correctly, therefore all the signals saved in the files are classified as EEG. We need to redefine this manually to get correct groups of sensors, we want to have only real EEG electrodes in the "EEG" category and put everything that we are not going to use a "MISC" category.
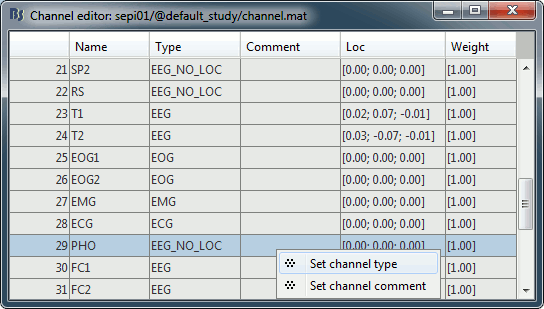
Right-click on the channel file > Edit channel file:
- Note that the EOG, EMG and ECG channels already have a different type
- Select all the other non-EEG channels: SP1, SP2, RS, PHO, DELR, DELL, QR, QL
Right-click in the window > Set channel type: type MISC
- Run the automatic alignment manually.
Mark and review spikes
- How was it done initially?
- How would we do it in Brainstorm?
- Import 177 spikes:
[-1, +1] seconds around the spike events (???)
- Remove DC offset based on the whole trial (-1s to +1s) (???)
Source analysis
- Compute head model: OpenMEEG or 3-shell sphere (???)
Noise covariance matrix: [-1000, -800] ms from all the imported trials (???)
- Source model: (???)
Moving dipoles
Illustrate John/Beth's tools for calculating and displaying dipoles.
Tools to be developed for this tutorial
- Set the time resolution of the display figures in seconds per centimeter (s/cm): the length of the page changes automatically with the size of the window.
- Flip +/- the Y axis (already done: check the brainstorm toolbar)
- Refine a bit the display of the dipoles
- Flexible montage interface (is this useful here ???)