|
Size: 1533
Comment:
|
Size: 23656
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| ## page was renamed from Epileptogenicity | |
| Line 3: | Line 2: |
| '''[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE] ''' |
|
| Line 9: | Line 10: |
| ''Warning: The same tools can be used for processing and displaying ECOG, but this hasn't been debugged properly yet. Please let us know how to improve the ECOG support.'' |
|
| Line 13: | Line 16: |
| Line 16: | Line 17: |
| How to get the example dataset: * Download zip: https://gin11-git.ujf-grenoble.fr/ftract_dev/ImaGIN_datasets/tree/master/tutorial_epimap * Unzip on your computer, rename to "tutorial_epimap" Files included in this package: * anat/MRI/3DT1*_deface.nii: Subject MRI before and after implantation, de-identified with FreeSurfer's mri_deface * anat/MRI/brainvisa: Cortical surface extracted with BrainVISA 4.5 * anat/implantation/*.txt: Positions of the sEEG contacts in various formats (MNI coordinates or patient coordinates) * seeg/SZ*.TRC: Seizure recordings in Micromed format, one seizure per file |
* '''Requirements''': You have already followed all the introduction tutorials and you have a working copy of Brainstorm installed on your computer. * Go to the [[http://neuroimage.usc.edu/bst/download.php|Download]] page of this website, and download the file: '''tutorial_epimap.zip''' * Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder) * Start Brainstorm (Matlab scripts or stand-alone version). * Select the menu File > Create new protocol. Name it "'''TutorialEpimap'''" and select the options: * "'''No, use individual anatomy'''", * "'''No, use one channel file per acquisition run'''". == Import the anatomy == === Pre-implantation MRI === * Right-click on the TutorialEpimap folder > '''New subject''' > '''Subject01'''.<<BR>>Keep the default options you defined for the protocol. * Switch to the "anatomy" view of the protocol. * Right-click on the subject node > '''Import MRI''':<<BR>>Set the file format: "All MRI file (subject space)"<<BR>>Select the file: '''tutorial_epimap/anat/MRI/3DT1pre_deface.nii''' * Do you want to apply the transformation to the MRI file? '''YES''' * The MRI viewer opens automatically. Click on "[[http://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy#MNI_transformation|Click here to compute MNI transformation]]". It computes an affine transformation between the subject space and the MNI ICBM152 space, and sets default positions for all the anatomical landmarks.<<BR>> {{attachment:mni_transformation.gif}} * Click on [Save] to close the MRI viewer. === Post-implantation MRI === * The pre-implantation MRI will be used as the anatomical reference for this subject. We will now import a second scan done after the SEEG implantation, on which we can see the SEEG electrodes and contacts. In this dataset, the post-implantation volume is another T1 MRI scan (contacts hyposignal appear in black), but it is more commonly a CT scan (contacts hypersignal appear in white). * Right-click on the subject node > '''Import MRI''':<<BR>>Select the file: '''tutorial_epimap/anat/MRI/3DT1post_deface.nii''' * Do you want to apply the transformation to the MRI file? '''YES'''<<BR>>This will reorient the MRI in Brainstorm's standard orientation, so you can see the coronal/sagittal/axial views correctly oriented. * How to register the new volume? '''IGNORE'''<<BR>>The two volumes have already been coregistered with SPM. See the section [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Volume_coregistration|Volume coregistration]] for more details on this option. * Reslice the volume? '''YES'''<<BR>>This will rewrite the volume with the orientation and resolution of the pre-implantation MRI, so that the two volumes can be overlaid in the MRI viewer. You may answer no to this question when the resolution of the pre-implantation MRI is very low and/or when you want to edit the positions of the fiducials in the original post-implantation volume (CT or MRI). * The MRI viewer opens automatically, showing the post-implantation volume as an colored layer on top of the previous volume. Adjust the transparency and amplitude threshold of this layer in the section Data options in the Surface tab, adjust its colormap with the popup menu of the figure. Use this display to validate that the coregistration of the two volume is correct, all the parts of the head must align well. <<BR>><<BR>> {{attachment:anat_overlay.gif||width="591",height="277"}} === Generate default surfaces === * Right-click on the pre-implantation MRI > '''SPM canonical surfaces'''. * Leave the default option selected (20484). This represents the resolution of SPM template surface used in this process. The higher the better, but it will slow down significantly the computation of the epileptogenicity maps. * These surfaces will be used later, in the computation of the epileptogenicity maps. Read the advanced sections of this page for information on [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Importing_realistic_surfaces|how to use realistic surfaces]] from BrainVISA or FreeSurfer.<<BR>><<BR>> {{attachment:anat_canonical.gif||width="598",height="161"}} == Access the recordings == === Link the recordings === * Switch to the "functional data" view (2nd button, on top of the database explorer). * Right-click on the subject folder > '''Review raw file''': * Select the file format: "'''SEEG: Deltamed/Micromed/...'''" * Select all the SEEG recordings: '''tutorial_epimap/seeg/*.TRC''' * The new files "Link to raw file" let you access directly the contents of the original SEEG files. The menu "Review raw file" does not actually copy any data to the database. [[http://neuroimage.usc.edu/brainstorm/Tutorials/ChannelFile#Review_vs_Import|More information]]. <<BR>><<BR>> {{attachment:import_link.gif||width="634",height="202"}} === Import the contacts positions === In order to generate epileptogenicity maps, we need accurate 3D positions for the contacts of the depth electrodes. Placing the contacts requires a good understanding of the implantation scheme reported by the neurosurgeon, and some skills in reading MRI scans. To make this tutorial easier to reproduce and follow, we distribute the positions of the contacts saved in a text file (folder /anat/implantation). For instructions to place the SEEG contacts using Brainstorm, read the advanced section [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Editing_the_contacts_positions|Editing the contacts positions]]. * The channel files "Micromed channels" contain the name of the channels, but not their positions. We need to import or edit separately the positions of the SEEG contacts. * Click on the [+] next to the three SZ folders, select all the channel files simultaneously. * Right-click one channel file > '''Add EEG positions''' > '''Import from file''': * Select the file format: "'''EEG: ASCII: Name, XYZ (*.*)'''" * Select the file: '''tutorial_epimap/anat/implantation/elec_pos_patient.txt '''<<BR>><<BR>> {{attachment:import_pos_file.gif||width="585",height="178"}} * Select a scaling factor: '''0.1''' (keep the default selection)<<BR>>The positions in this text file are in millimeters, while the expected unit for "EEG: ASCII" is the centimeter. This option means that the values will be interpreted as 0.1*cm, ie. millimeters. <<BR>><<BR>> {{attachment:import_pos_factor.gif||width="407",height="111"}} * Apply MRI transformation? '''YES'''<<BR>>This will interpret the coordinates in the text file as positions in the referential defined by the voxel-to-world transformation (vox2ras) transformation available in the MRI, if available (in NIfTI format, this corresponds to matrices qform or sform). If you answer no here, it would load the positions as [[CoordinateSystems|Brainstorm SCS coordinates]] and try to realign them based on the NAS/LPA/RPA landmarks. <<BR>><<BR>> {{attachment:import_pos_transf.gif||width="345",height="98"}} * At the end you get a report indicating how many channels from the SEEG recordings were attributed a new 3D position. The channels are matched by name: the position file you import must include the labels of the electrodes and they must be named exactly in the the same way as in your recordings. * The type of the channels for which a position was not found is set to '''EEG_NO_LOC''', so they are ignored when you process the SEEG channels. In this example dataset, the channels that are not found are "fz" and "cz", which are indeed not SEEG contacts. * You should always validate that the type of all the channels has been detected correctly. Right-click on a channel file > '''Edit channel file'''.<<BR>><<BR>> {{attachment:import_pos_edit.gif||width="595",height="120"}} * '''MNI coordinates''': Note that you can also import contact positions in MNI coordinates. In this example dataset includes the positions, they are available in the file "elec_pos_patient.txt". To import this file correctly, make sure you select a file format that explicitely mentions MNI coordinates. In this example, it would be: "EEG: ASCII: Name, XYZ'''_MNI''' (*.*)". * If you don't have the positions for the SEEG contacts, or if they don't look correctly aligned, you can directly place them in the MRI viewer. See the section [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epileptogenicity#Editing_the_contacts_positions|Editing the contacts positions]]. == Display the depth electrodes == === 3D figures === * Right-click on the channel file > '''Display sensors''' > Explore all the options available. {{attachment:display_menu.gif}} * You can render the SEEG depth electrodes in 3D together with the subject surfaces, pre- or post-implantation volumes. You can add more anatomy elements to the figure with the button "Add a surface" at the top-right of the Surface tab. For more help: [[http://neuroimage.usc.edu/brainstorm/Tutorials/ExploreAnatomy#MRI_in_3D|Display the anatomy]]. <<BR>><<BR>> {{attachment:display_3d.gif||width="507",height="273"}} * Click on contact to select it, right-click on the figure to get its name: {{attachment:display_selectchan.gif||width="408",height="247"}} === MRI Viewer === * You can also display the contacts in the MRI viewer, on top of the the pre- or post-implantation volumes. By default, the electrode is displayed in a slice if there is a contact of attached to this electrode in the slice.<<BR>><<BR>> {{attachment:display_mri.gif||width="542",height="422"}} * To display all the electrodes, select the option "'''MIP: Functional'''". For a glass-brain view, select at the same time the option "'''MIP: Anatomy'''". <<BR>><<BR>> {{attachment:display_mri_mip.gif||width="621",height="508"}} * Zoom in/out with the buttons or the associated shortcuts (Ctrl+Scroll or +/-) and explore the volume with (Shift+)x/y/z. Additional display options are available in the popup menu for this figure. All the shortcuts listed in [[http://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy#Using_the_MRI_Viewer|this tutorial]]. * You can '''edit the position of a contact''' directly by clicking on its dot in one of the slice and moving it around. Modifications are not saved immediately and can be cancelled when you close the window. Alternatively, you can right-click on the figure > '''Set electrode position'''. === Panel iEEG === * When opening SEEG/ECOG recordings, the panel iEEG is added to the Brainstorm window. You can use it to edit the display properties of the depth electrodes. In this interface, "'''electrode'''" refers to entire depth electrode implanted in the head of the patient while "'''contact'''" refers to recording sites on the electrode. There are multiple contacts on an electrode, and one contact corresponds to one channel of data in the channel file and the recordings. <<BR>><<BR>> {{attachment:panel_ieeg.gif||width="611",height="319"}} * SEEG depth electrodes are graphical objects, they are defined independently from the SEEG contacts available in the channel file and recordings. A contact/channel is associated to a depth electrode using the '''Group''' property of the channel, accessible with a right-click on the channel file > '''Edit channel file'''. * In this example, the depth electrodes available in the panel iEEG have been detected automatically based on the names and positions of the data channels. In the convention used here, the contact names must start with one or more letters (the name of the electrode) followed by a number representing the index of the contact on the electrode. Contact #1 is at the tip of the electrode, and is therefore the deeper contact of the electrode. If the convention used in your recordings is different, you may have to edit the electrodes properties in order to get them displayed correctly. * Buttons in the toolbar: * '''Add an electrode''': Adds an entry for a new depth electrode. The new electrode will not be displayed until you set its properties and position. This will not add or remove SEEG contacts or channels of data. * '''Remove selected electrodes''': Deletes a depth electrode from the list, but does not modify the list of SEEG contacts or channels of data. * '''Set color for selected electrodes''': Self explanatory. * '''Show/hide selected electrodes''': To hide an electrode in the 3D figures and MRI viewers, select it in the list then click on this button. Select all the electrodes with the standard shortcut Ctrl+A. * '''Display contacts as''': Depth electrodes / Spheres. * '''Contacts > Set default positions''': For each of the selected electrodes, the current positions of the SEEG contacts are discarded and replaced with the default positions of the contacts on the electrode. The properties used for setting the position of the contacts are the contact spacing, the tip of the electrode and the entry point in the skull. Contact #i is placed along the electrode at (i-1)*contact_spacing millimeters for the tip of the electrode. * '''Contacts > Project on electrode''': For each of the selected electrodes, the contacts are projected orthogonally on the electrode. This menu can be useful for aligning contacts that were marked one by one. * '''Contacts > Save modifications''': Save the current modifications to the channel file. Otherwise, the modifications are saved only when you close the figure (dialog box "Save modifications to channel file?") * '''Contacts > Export contacts positions''': Save the 3D positions of the SEEG contacts in the current channel file in one of the file formats supported by Brainstorm. * Electrode properties: ''Properties that have an impact on the position of the contacts''.<<BR>> * If you edit the properties, the modifications will apply to all the selected electrodes in the current channel file. Check what is selected before making changes. * '''Type''': SEEG/ECOG * '''Model''': List of electrode models. If you select an entry in this menu, it will copy the default properties for this model to the selected electrodes. If you are using electrodes that are not in this list, please post on the specification of your devices on the user forum and we will add them to this list. * '''Number of contacts''': Number of recording sites on the electrode. By default, this is set for SEEG to the maximum index found in a group of contacts. Example: electrode s' is associated to channels s'1, s'2, s'3, s'10, s'11, s'12 => detected number of contacts is 12. * '''Contact spacing''': Distance between the centers of two consecutive contacts in the electrode. In this example dataset, the default value corresponds to the average distance observed between pairs of adjacent contacts. * '''Set tip''': Select one electrode in the list, then move the cursor of the MRI viewer to the tip of the selected electrode (center of the first contact), and finally click on [Set tip]. * '''Set skull entry''': Select one electrode in the list, then move the cursor of the MRI viewer to the point where the depth electrode enters the skull (or the center of the screw), and finally click on [Set skull entry]. This position does not correspond to any contact, it is used only to estimate the direction of the depth electrode. * Display options: Properties that only affect the way the electrodes are renderer graphically. * '''Contact length''': Defines the length along the electrode axis of the yellow cylinder that represents the contacts, or the diameter of the sphere when the electrodes are not rendered. * '''Contact diameter''': Diameter of the yellow cylinders representing the contacts. By default, this value is slightly larger than the electrode diameter so that it is rendered correctly. If you use the same value as the electrode diameter, the contacts might not be visible. * '''Electrode diameter''': Diameter of the cylinder representing the SEEG depth electrode. * '''Electrode length''': Length of the cylinder representing the SEEG depth electrode. == Display the SEEG recordings == === SEEG time series === * Right-click on any "Link to raw file" > '''SEEG '''> '''Display time series'''.<<BR>><<BR>> {{attachment:display_ts.gif||width="637",height="276"}} * For instructions on how to use this viewer, please refer to these tutorials: * [[Tutorials/ReviewRaw|Review continuous recordings]] * [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy#Review_EEG_recordings|EEG and epilepsy: Review EEG recordings]] * Typical display configuration includes: * Set the page duration tp 20s or 30s: Record tab > Page settings. * Change the display mode to "columns": Record tab > Toolbar > First button. * Change the montage (bipolar/monopolar): Record tab > Note all the default montages available for SEEG: you can display all the contacts at once, or each electrode individually. * Set visualization filters: Filter tab. * Disable the automatic scaling when changing page: Button [AS] in the time series figure. * Adjust the amplitude scale: Right-click+move up/down, or buttons [^] and [v]. * Set the resolution precisely: Right-click on the figure > Figure > Set axes resolution. * With the menus "'''3D electrodes'''" you can visualize the SEEG values at the current time point. Move the time cursor and it will update the values represented on the SEEG contacts. <<BR>><<BR>> {{attachment:display_3delectrodes.gif||width="640",height="299"}} === Interpolate on the anatomy === * From the same SEEG display menu, you can also project the SEEG values on the MRI or the cortex. You can edit the colormap for the displayed value [[http://neuroimage.usc.edu/brainstorm/Tutorials/Colormaps|like any other colormap]]. * For the MRI, all the voxels in the neighborhood of a contact are attributed the value associated with this contact. For the surface, the values of the vertices are interpolated from the nearest SEEG contacts (the magnitude decreases with the distance to the contact). {{attachment:display_interp_anat.gif||width="640",height="263"}} == Bad channels == We will now to review the recordings for the three seizures recorded for this subject: SZ1, SZ2, SZ3. All the following steps are illustrated only for '''SZ1''' but need to be reproduced for the other files. * Double-click on '''SZ1/Link to raw file''' to open the SEEG recordings for the first seizure. * We need to mark the bad channels using a "'''monopolar'''" montage (common reference). If you are currently reviewing the recordings with a bipolar montage, swith it back to "All channels", using the menu in the toolbar of the record tab or with the popup menu of the time series figure. Otherwise, marking a signal as bad (eg. "v'2-v'1") would actually mark two channels as bad ("v'1" and "v'2"), while only one contact ("v'1") is not recording properly. * Click on the channels that you consider as bad to select them. For SZ1, select '''v'1''' and '''f'1'''. * Press the '''delete''' key, or use the popup menu Channels > '''Mark selected as bad'''. <<BR>><<BR>> {{attachment:mark_bad.gif||width="629",height="296"}} * Close the figure and repeat for the two other files. * Bad channels for the three seizures: * SZ1: '''v'1''', '''f'1''' * SZ2: '''v'1''' * SZ3: '''o'1''' == Edit event markers == === Seizure onset === Two events are already available in this file: "Part1" only indicates the beginning of the file, and "Seizure" indicates approximately the onset of the first seizure. This marking was done for clinical use, to jump quickly to the page of interest, but here we need a more accurate marker. We will create a new marker "Onset" where we observe the beginning of the fast gamma activity, which is the main feature used computation of the epileptogenicity maps is based on. * Double-click on '''SZ1/Link to raw file''' to open the SEEG recordings for the first seizure, or do this immediately after marking the bad channels. * Switch to a '''bipolar montage''': bipolar-1 is sparse (eg. g2-g1, g4-3), bipolar-2 is exhaustive (eg. g2-g1, g3-g2, g4-3), depending on how dense you want your figure to be. * In the Record tab, click on the event group "'''Seizure'''", then on the event timing. This will jump quickly to the page on which we will place our Onset marker. * Zoom around the '''g'''' contacts (using the buttons on the right of the figure or the corresponding shortcuts, indicated if you leave your mouse over the buttons for a second), or select the g' electrode in the '''montage menu'''. * If needed, zoom locally in time in the current page (mouse wheel) and change the amplitude scale (right-click+mouse up/down). * Create an event group "Onset": Record tab > Events > '''Add group > "Onset"''', or select it. * Place the time cursor at '''120.800s''', add an Onset marker at this time (press the key "E", or use the popup menu Add/delete event). <<BR>><<BR>> {{attachment:mark_onset.gif}} * If you are marking a lot of events, you might be interest in using [[http://neuroimage.usc.edu/brainstorm/Tutorials/EventMarkers#Custom_shortcuts|custom shortcuts]]. === Baseline === In the following sections, we will compute a time-frequency decomposition of the seizure and normalize it with respect to a baseline level. We will identify and import this baseline now. The baseline could be a segment of 5s to 30s of recordings, typically before the seizure, that contains as little interictal epileptiform activity as possible. The duration of the baseline influences the computation time for the following steps of this tutorial. We will work here with short segments of 5s for illustration purposes, between 72.8s and 77.8s. Use longer segments for more accurate results. . . . == Import epochs of interest == == Time-frequency analysis == == Epileptogenicity maps == === Volume === === Surface === <<TAG(Advanced)>> == Guidelines panel == Easy way for reproducing all these steps <<TAG(Advanced)>> == On the hard drive == IntraElectrodes structure <<TAG(Advanced)>> == Volume coregistration == <<TAG(Advanced)>> == Editing the contacts positions == <<TAG(Advanced)>> == Creating a new implantation file == Same as previous section, but not starting from an existing of contacts. <<TAG(Advanced)>> == Importing realistic surfaces == == TODO == * Fix all the links in this page * Proofread and check spelling |
SEEG epileptogencity maps
[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE]
Authors: Francois Tadel, Olivier David.
This tutorial introduces some concepts that are specific to the management of SEEG recordings in the Brainstorm environment, and explains how to compute maps of epileptogenicity from ictal recordings. It is based on a clinical case from the Grenoble University Hospital, France.
Note that the operations used here are not detailed, the goal of this tutorial is not to introduce Brainstorm to new users. For in-depth explanations of the interface and theoretical foundations, please refer to the introduction tutorials.
Warning: The same tools can be used for processing and displaying ECOG, but this hasn't been debugged properly yet. Please let us know how to improve the ECOG support.
Contents
- Dataset description
- Download and installation
- Import the anatomy
- Access the recordings
- Display the depth electrodes
- Display the SEEG recordings
- Bad channels
- Edit event markers
- Import epochs of interest
- Time-frequency analysis
- Epileptogenicity maps
- Guidelines panel
- On the hard drive
- Volume coregistration
- Editing the contacts positions
- Creating a new implantation file
- Importing realistic surfaces
- TODO
Dataset description
License
This tutorial dataset (EEG and MRI data) remains property of the Grenoble University Hospital, France. Its use and transfer outside the ImaGIN and Brainstorm tutorials, e.g. for research purposes, is prohibited without written consent. For questions, please contact Olivier David, PhD ( olivier.david@inserm.fr ).
Clinical description
This dataset includes recordings for a patient that was not reported in the above article, but are part of the same study. The patient presents a focal epilepsy of the left temporo-occipital junction, MRI-negative, and was implanted in the surrounding areas. The subfolder "seeg" contains the recordings of three seizures, all of them showing a propagation of high-frequency oscillations from the lesion towards the temporal lobe, bilaterally.
SEEG recordings
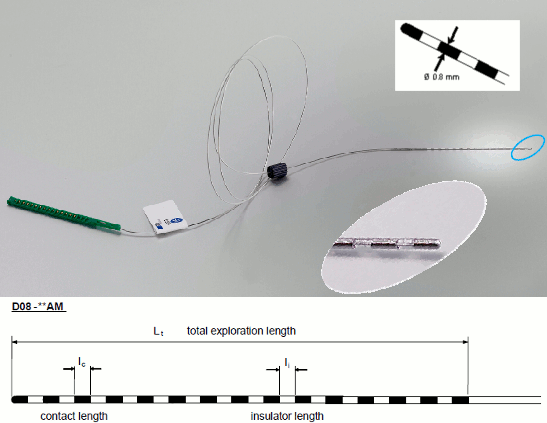
The depth electrodes used in this example dataset are DIXI D08-**AM Microdeep electrodes, with the following specifications:
- Diameter: 0.8 mm
- Contact length: 2 mm
- Insulator length: 1.5 mm
- Distance between the center of two contacts: 3.5 mm
- Between 8 and 18 contacts on each electrode
Files
The dataset we distribute with this tutorial follows the Brain Imaging Data Structure (BIDS) standard for neuroimaging data organization. This specification was first established for MRI and fMRI (Gorgolewski, 2016) and then refined with an extension dedicated to iEEG (Holdgraf, 2019). The files that will be imported in this tutorial are the following:
tutorial_epimap_bids/
derivatives/: Everything that cannot be considered as raw data
brainvisa/sub-01_ses-pre/: Result of the BrainVISA 4.5 segmentation for the pre-implantation T1 MRI.
sub-01/: Raw data for subject 01
ses-preimp/: Imaging exams performed before the implantation of the sEEG.
anat/sub-01_ses-preimp_T1w.nii.gz: T1-weighted MRI pre-implantation
ses-postimp/: Exams performed with the sEEG devices implanted.
anat/sub-01_ses-postimp_T1w.nii.gz: T1-weighted MRI post-implantation
ieeg/..._task-seizure_run-0*_ieeg.eeg: Three seizure recordings in BrainVision file format, one seizure per file (with the header files .vhdr and .vmrk)
ieeg/..._space-IXI549Space_electrodes.tsv: Position of the contacts in MNI space (SPM12 Segment non-linear normalization)
ieeg/..._space-ScanRAS_electrodes.tsv: Position of the contacts in world coordinates, relative to the post-implantation T1 MRI.
All the anatomical images have been de-identified with mri_deface from FreeSurfer 6.
References
The acquisition methodology is described in the following articles:
David O, Blauwblomme T, Job AS, Chabardès S, Hoffmann D, Minotti L, Kahane P, Imaging the seizure onset zone with stereo-electroencephalography, Brain. 2011 Oct;134(10):2898-911
Lamarche F, Job AS, Deman P, Bhattacharjee M, Hoffmann D, Gallazzini-Crépin C, Bouvard S, Minotti L, Kahane P, David O, Correlation of FDG-PET hypometabolism and SEEG epileptogenicity mapping in patients with drug-resistant focal epilepsy, Epilepsia. 2016 Dec; 57(12):2045–2055
Download and installation
Requirements: You have already followed all the introduction tutorials and you have a working copy of Brainstorm installed on your computer.
Go to the Download page of this website, and download the file: tutorial_epimap.zip
- Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder)
- Start Brainstorm (Matlab scripts or stand-alone version).
Select the menu File > Create new protocol. Name it "TutorialEpimap" and select the options:
"No, use individual anatomy",
"No, use one channel file per acquisition run".
Import the anatomy
Pre-implantation MRI
Right-click on the TutorialEpimap folder > New subject > Subject01.
Keep the default options you defined for the protocol.- Switch to the "anatomy" view of the protocol.
Right-click on the subject node > Import MRI:
Set the file format: "All MRI file (subject space)"
Select the file: tutorial_epimap/anat/MRI/3DT1pre_deface.niiDo you want to apply the transformation to the MRI file? YES
The MRI viewer opens automatically. Click on "Click here to compute MNI transformation". It computes an affine transformation between the subject space and the MNI ICBM152 space, and sets default positions for all the anatomical landmarks.
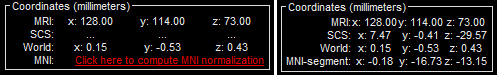
- Click on [Save] to close the MRI viewer.
Post-implantation MRI
- The pre-implantation MRI will be used as the anatomical reference for this subject. We will now import a second scan done after the SEEG implantation, on which we can see the SEEG electrodes and contacts. In this dataset, the post-implantation volume is another T1 MRI scan (contacts hyposignal appear in black), but it is more commonly a CT scan (contacts hypersignal appear in white).
Right-click on the subject node > Import MRI:
Select the file: tutorial_epimap/anat/MRI/3DT1post_deface.niiDo you want to apply the transformation to the MRI file? YES
This will reorient the MRI in Brainstorm's standard orientation, so you can see the coronal/sagittal/axial views correctly oriented.How to register the new volume? IGNORE
The two volumes have already been coregistered with SPM. See the section Volume coregistration for more details on this option.Reslice the volume? YES
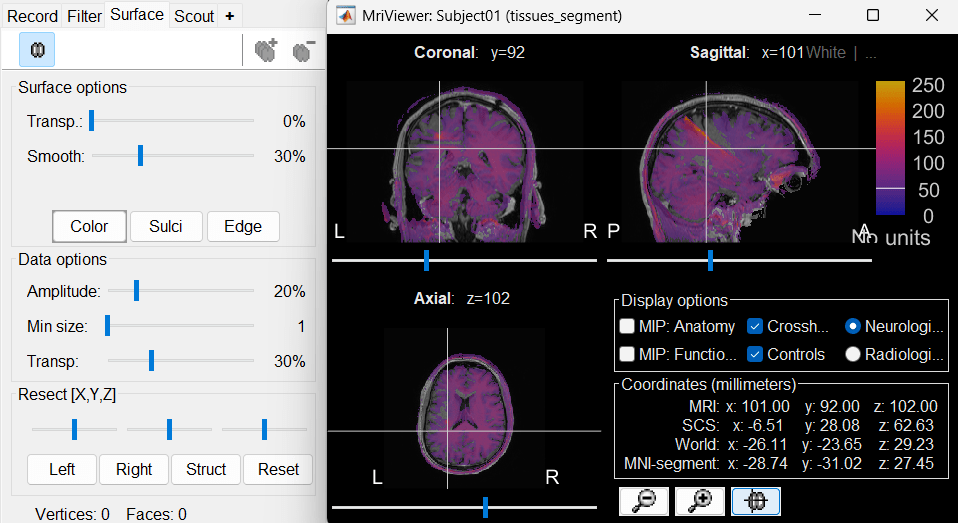
This will rewrite the volume with the orientation and resolution of the pre-implantation MRI, so that the two volumes can be overlaid in the MRI viewer. You may answer no to this question when the resolution of the pre-implantation MRI is very low and/or when you want to edit the positions of the fiducials in the original post-implantation volume (CT or MRI).The MRI viewer opens automatically, showing the post-implantation volume as an colored layer on top of the previous volume. Adjust the transparency and amplitude threshold of this layer in the section Data options in the Surface tab, adjust its colormap with the popup menu of the figure. Use this display to validate that the coregistration of the two volume is correct, all the parts of the head must align well.
Generate default surfaces
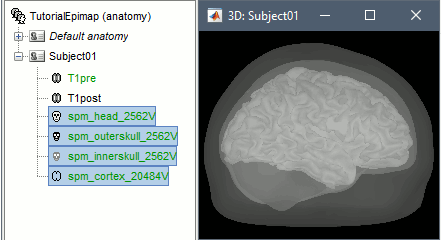
Right-click on the pre-implantation MRI > SPM canonical surfaces.
- Leave the default option selected (20484). This represents the resolution of SPM template surface used in this process. The higher the better, but it will slow down significantly the computation of the epileptogenicity maps.
These surfaces will be used later, in the computation of the epileptogenicity maps. Read the advanced sections of this page for information on how to use realistic surfaces from BrainVISA or FreeSurfer.
Access the recordings
Link the recordings
- Switch to the "functional data" view (2nd button, on top of the database explorer).
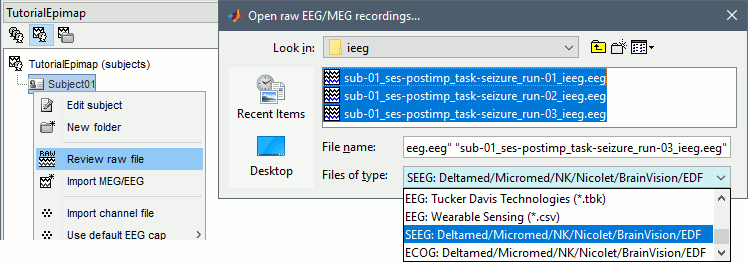
Right-click on the subject folder > Review raw file:
Select the file format: "SEEG: Deltamed/Micromed/..."
Select all the SEEG recordings: tutorial_epimap/seeg/*.TRC
The new files "Link to raw file" let you access directly the contents of the original SEEG files. The menu "Review raw file" does not actually copy any data to the database. More information.
Import the contacts positions
In order to generate epileptogenicity maps, we need accurate 3D positions for the contacts of the depth electrodes. Placing the contacts requires a good understanding of the implantation scheme reported by the neurosurgeon, and some skills in reading MRI scans. To make this tutorial easier to reproduce and follow, we distribute the positions of the contacts saved in a text file (folder /anat/implantation). For instructions to place the SEEG contacts using Brainstorm, read the advanced section Editing the contacts positions.
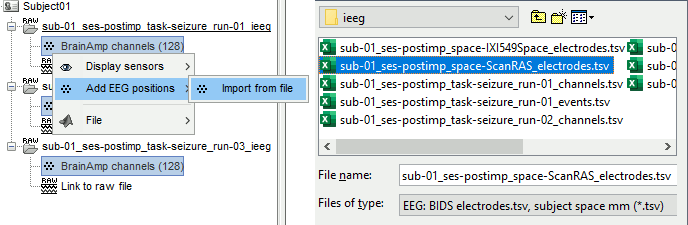
- The channel files "Micromed channels" contain the name of the channels, but not their positions. We need to import or edit separately the positions of the SEEG contacts.
- Click on the [+] next to the three SZ folders, select all the channel files simultaneously.
Right-click one channel file > Add EEG positions > Import from file:
Select the file format: "EEG: ASCII: Name, XYZ (*.*)"
Select the file: tutorial_epimap/anat/implantation/elec_pos_patient.txt
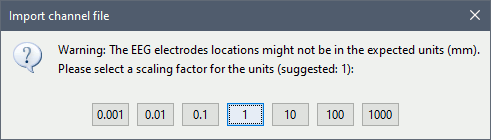
Select a scaling factor: 0.1 (keep the default selection)
The positions in this text file are in millimeters, while the expected unit for "EEG: ASCII" is the centimeter. This option means that the values will be interpreted as 0.1*cm, ie. millimeters.
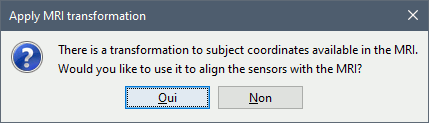
Apply MRI transformation? YES
This will interpret the coordinates in the text file as positions in the referential defined by the voxel-to-world transformation (vox2ras) transformation available in the MRI, if available (in NIfTI format, this corresponds to matrices qform or sform). If you answer no here, it would load the positions as Brainstorm SCS coordinates and try to realign them based on the NAS/LPA/RPA landmarks.
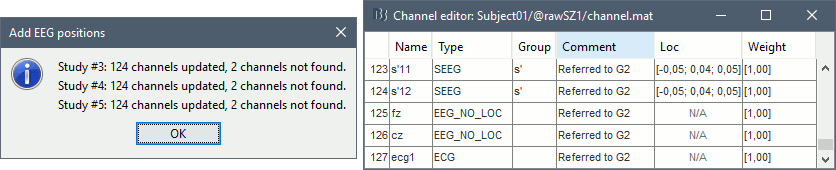
- At the end you get a report indicating how many channels from the SEEG recordings were attributed a new 3D position. The channels are matched by name: the position file you import must include the labels of the electrodes and they must be named exactly in the the same way as in your recordings.
The type of the channels for which a position was not found is set to EEG_NO_LOC, so they are ignored when you process the SEEG channels. In this example dataset, the channels that are not found are "fz" and "cz", which are indeed not SEEG contacts.
You should always validate that the type of all the channels has been detected correctly. Right-click on a channel file > Edit channel file.
MNI coordinates: Note that you can also import contact positions in MNI coordinates. In this example dataset includes the positions, they are available in the file "elec_pos_patient.txt". To import this file correctly, make sure you select a file format that explicitely mentions MNI coordinates. In this example, it would be: "EEG: ASCII: Name, XYZ_MNI (*.*)".
If you don't have the positions for the SEEG contacts, or if they don't look correctly aligned, you can directly place them in the MRI viewer. See the section Editing the contacts positions.
Display the depth electrodes
3D figures
Right-click on the channel file > Display sensors > Explore all the options available.
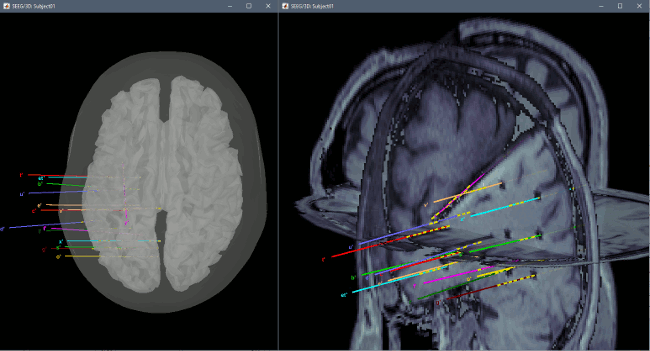
You can render the SEEG depth electrodes in 3D together with the subject surfaces, pre- or post-implantation volumes. You can add more anatomy elements to the figure with the button "Add a surface" at the top-right of the Surface tab. For more help: Display the anatomy.
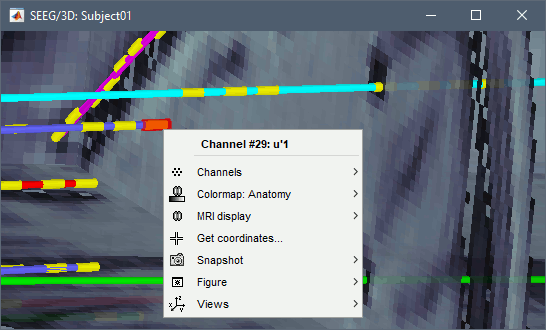
Click on contact to select it, right-click on the figure to get its name:
MRI Viewer
You can also display the contacts in the MRI viewer, on top of the the pre- or post-implantation volumes. By default, the electrode is displayed in a slice if there is a contact of attached to this electrode in the slice.
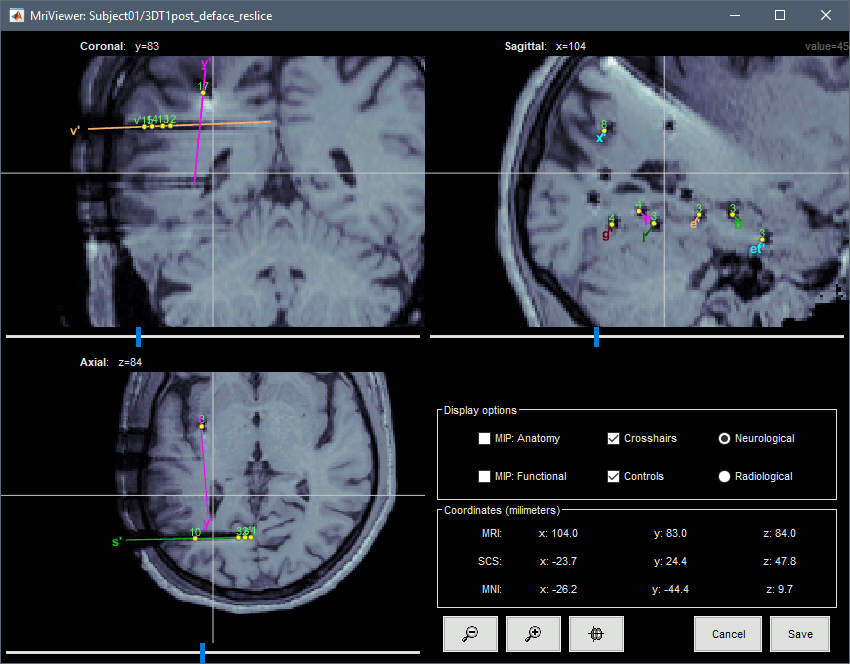
To display all the electrodes, select the option "MIP: Functional". For a glass-brain view, select at the same time the option "MIP: Anatomy".
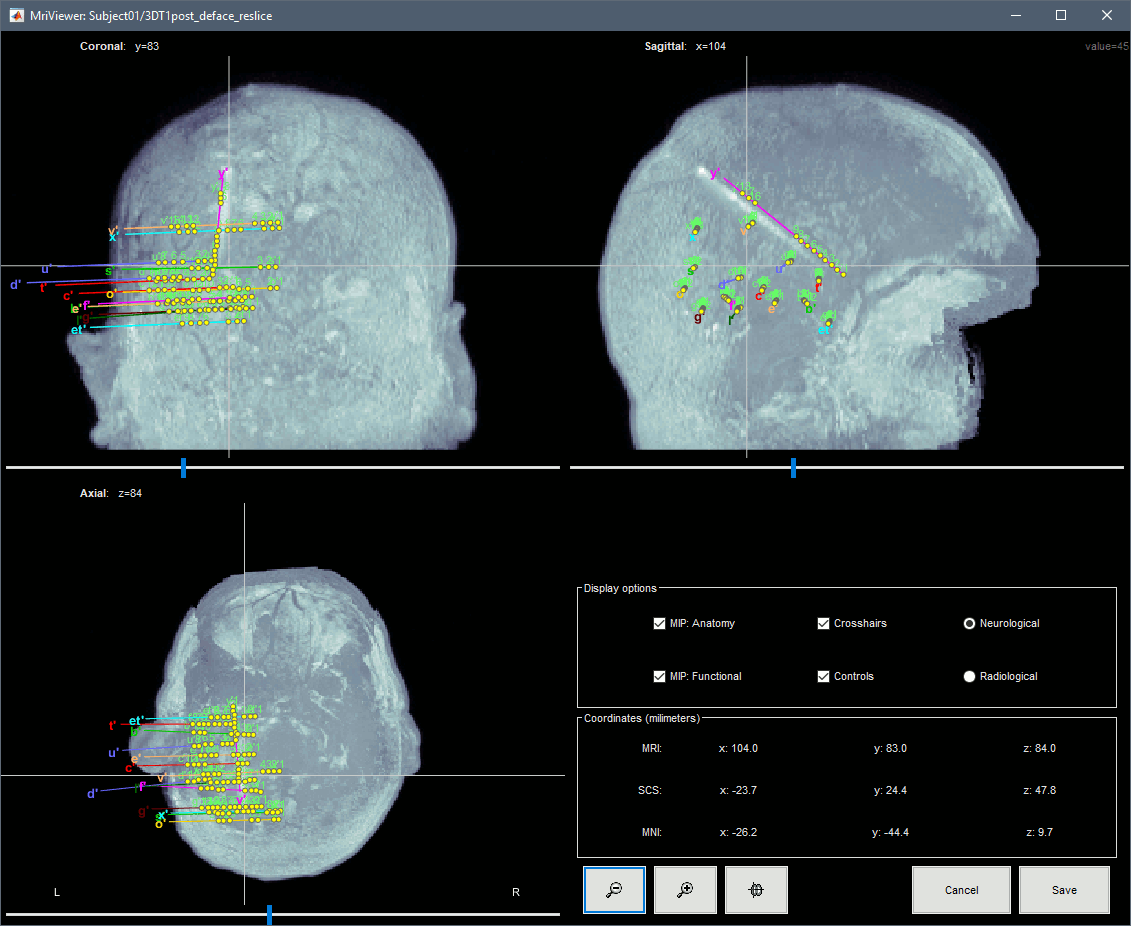
Zoom in/out with the buttons or the associated shortcuts (Ctrl+Scroll or +/-) and explore the volume with (Shift+)x/y/z. Additional display options are available in the popup menu for this figure. All the shortcuts listed in this tutorial.
You can edit the position of a contact directly by clicking on its dot in one of the slice and moving it around. Modifications are not saved immediately and can be cancelled when you close the window. Alternatively, you can right-click on the figure > Set electrode position.
Panel iEEG
When opening SEEG/ECOG recordings, the panel iEEG is added to the Brainstorm window. You can use it to edit the display properties of the depth electrodes. In this interface, "electrode" refers to entire depth electrode implanted in the head of the patient while "contact" refers to recording sites on the electrode. There are multiple contacts on an electrode, and one contact corresponds to one channel of data in the channel file and the recordings.
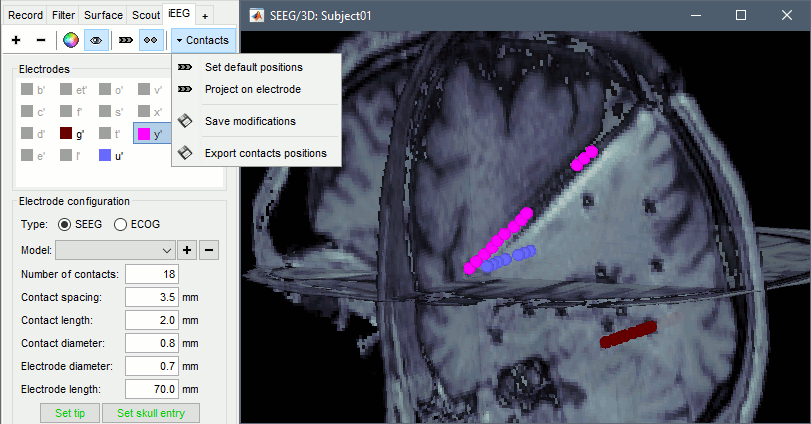
SEEG depth electrodes are graphical objects, they are defined independently from the SEEG contacts available in the channel file and recordings. A contact/channel is associated to a depth electrode using the Group property of the channel, accessible with a right-click on the channel file > Edit channel file.
- In this example, the depth electrodes available in the panel iEEG have been detected automatically based on the names and positions of the data channels. In the convention used here, the contact names must start with one or more letters (the name of the electrode) followed by a number representing the index of the contact on the electrode. Contact #1 is at the tip of the electrode, and is therefore the deeper contact of the electrode. If the convention used in your recordings is different, you may have to edit the electrodes properties in order to get them displayed correctly.
- Buttons in the toolbar:
Add an electrode: Adds an entry for a new depth electrode. The new electrode will not be displayed until you set its properties and position. This will not add or remove SEEG contacts or channels of data.
Remove selected electrodes: Deletes a depth electrode from the list, but does not modify the list of SEEG contacts or channels of data.
Set color for selected electrodes: Self explanatory.
Show/hide selected electrodes: To hide an electrode in the 3D figures and MRI viewers, select it in the list then click on this button. Select all the electrodes with the standard shortcut Ctrl+A.
Display contacts as: Depth electrodes / Spheres.
Contacts > Set default positions: For each of the selected electrodes, the current positions of the SEEG contacts are discarded and replaced with the default positions of the contacts on the electrode. The properties used for setting the position of the contacts are the contact spacing, the tip of the electrode and the entry point in the skull. Contact #i is placed along the electrode at (i-1)*contact_spacing millimeters for the tip of the electrode.
Contacts > Project on electrode: For each of the selected electrodes, the contacts are projected orthogonally on the electrode. This menu can be useful for aligning contacts that were marked one by one.
Contacts > Save modifications: Save the current modifications to the channel file. Otherwise, the modifications are saved only when you close the figure (dialog box "Save modifications to channel file?")
Contacts > Export contacts positions: Save the 3D positions of the SEEG contacts in the current channel file in one of the file formats supported by Brainstorm.
Electrode properties: Properties that have an impact on the position of the contacts.
- If you edit the properties, the modifications will apply to all the selected electrodes in the current channel file. Check what is selected before making changes.
Type: SEEG/ECOG
Model: List of electrode models. If you select an entry in this menu, it will copy the default properties for this model to the selected electrodes. If you are using electrodes that are not in this list, please post on the specification of your devices on the user forum and we will add them to this list.
Number of contacts: Number of recording sites on the electrode. By default, this is set for SEEG to the maximum index found in a group of contacts. Example: electrode s' is associated to channels s'1, s'2, s'3, s'10, s'11, s'12 => detected number of contacts is 12.
Contact spacing: Distance between the centers of two consecutive contacts in the electrode. In this example dataset, the default value corresponds to the average distance observed between pairs of adjacent contacts.
Set tip: Select one electrode in the list, then move the cursor of the MRI viewer to the tip of the selected electrode (center of the first contact), and finally click on [Set tip].
Set skull entry: Select one electrode in the list, then move the cursor of the MRI viewer to the point where the depth electrode enters the skull (or the center of the screw), and finally click on [Set skull entry]. This position does not correspond to any contact, it is used only to estimate the direction of the depth electrode.
- Display options: Properties that only affect the way the electrodes are renderer graphically.
Contact length: Defines the length along the electrode axis of the yellow cylinder that represents the contacts, or the diameter of the sphere when the electrodes are not rendered.
Contact diameter: Diameter of the yellow cylinders representing the contacts. By default, this value is slightly larger than the electrode diameter so that it is rendered correctly. If you use the same value as the electrode diameter, the contacts might not be visible.
Electrode diameter: Diameter of the cylinder representing the SEEG depth electrode.
Electrode length: Length of the cylinder representing the SEEG depth electrode.
Display the SEEG recordings
SEEG time series
Right-click on any "Link to raw file" > SEEG > Display time series.
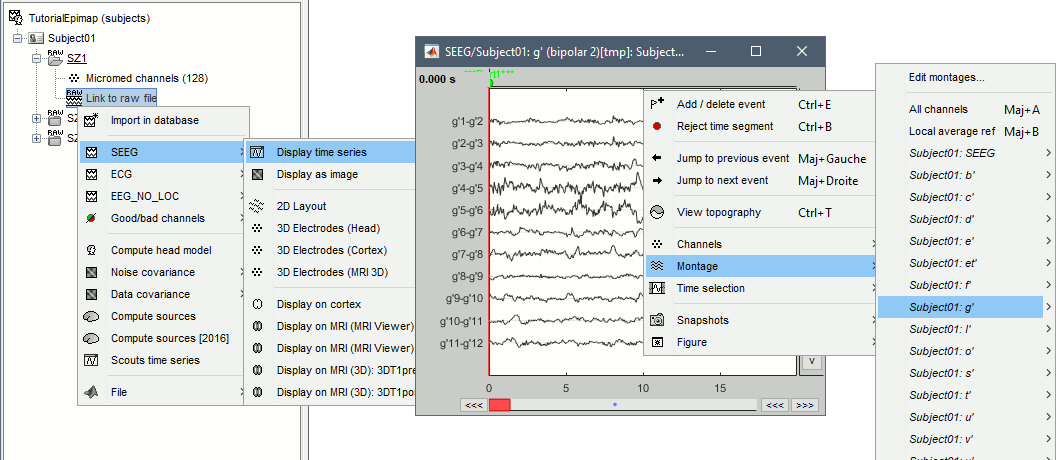
- For instructions on how to use this viewer, please refer to these tutorials:
- Typical display configuration includes:
Set the page duration tp 20s or 30s: Record tab > Page settings.
Change the display mode to "columns": Record tab > Toolbar > First button.
Change the montage (bipolar/monopolar): Record tab > Note all the default montages available for SEEG: you can display all the contacts at once, or each electrode individually.
- Set visualization filters: Filter tab.
- Disable the automatic scaling when changing page: Button [AS] in the time series figure.
- Adjust the amplitude scale: Right-click+move up/down, or buttons [^] and [v].
Set the resolution precisely: Right-click on the figure > Figure > Set axes resolution.
With the menus "3D electrodes" you can visualize the SEEG values at the current time point. Move the time cursor and it will update the values represented on the SEEG contacts.
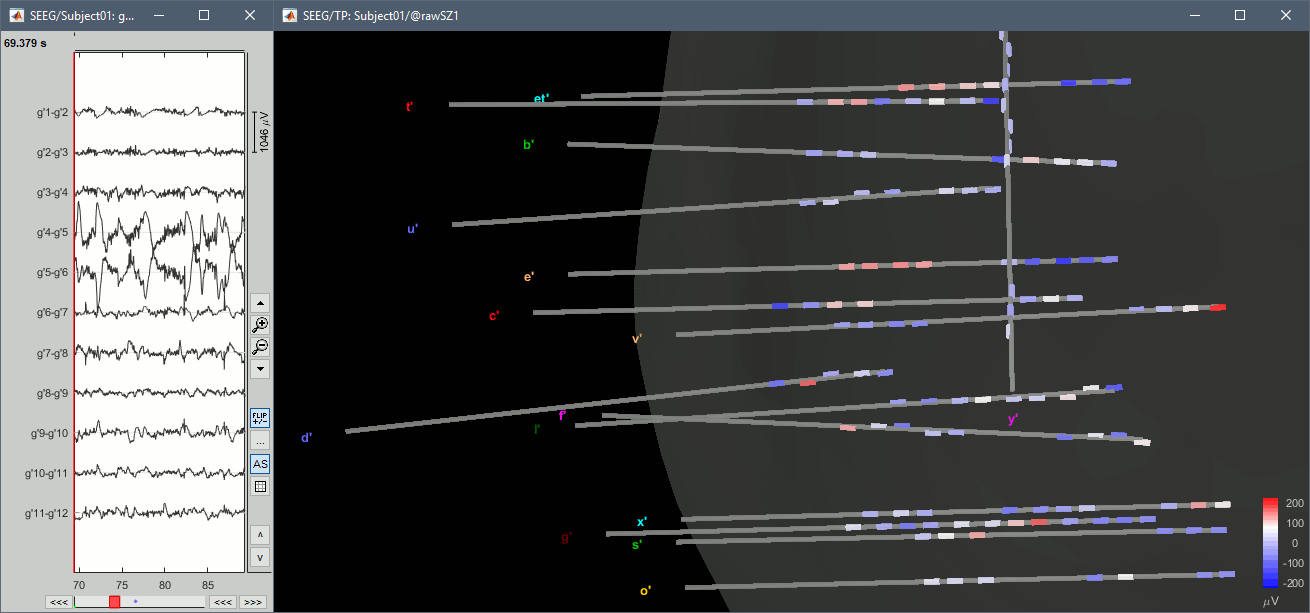
Interpolate on the anatomy
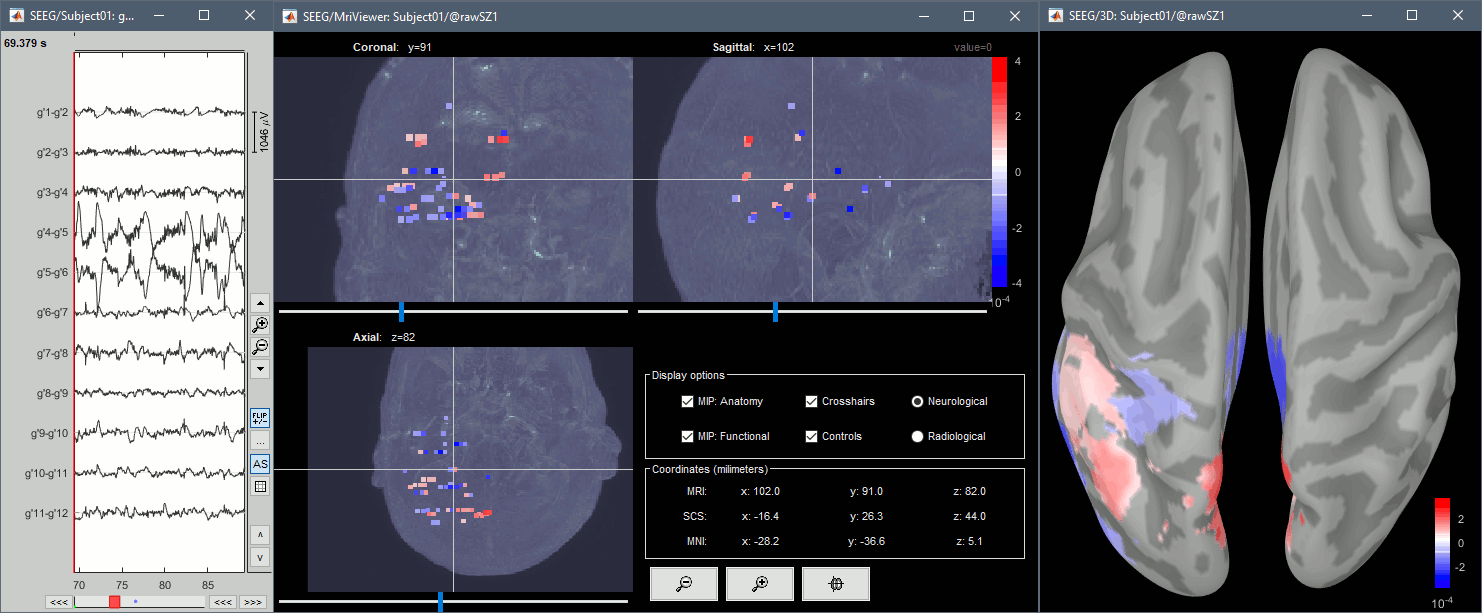
From the same SEEG display menu, you can also project the SEEG values on the MRI or the cortex. You can edit the colormap for the displayed value like any other colormap.
For the MRI, all the voxels in the neighborhood of a contact are attributed the value associated with this contact. For the surface, the values of the vertices are interpolated from the nearest SEEG contacts (the magnitude decreases with the distance to the contact).
Bad channels
We will now to review the recordings for the three seizures recorded for this subject: SZ1, SZ2, SZ3. All the following steps are illustrated only for SZ1 but need to be reproduced for the other files.
Double-click on SZ1/Link to raw file to open the SEEG recordings for the first seizure.
We need to mark the bad channels using a "monopolar" montage (common reference). If you are currently reviewing the recordings with a bipolar montage, swith it back to "All channels", using the menu in the toolbar of the record tab or with the popup menu of the time series figure. Otherwise, marking a signal as bad (eg. "v'2-v'1") would actually mark two channels as bad ("v'1" and "v'2"), while only one contact ("v'1") is not recording properly.
Click on the channels that you consider as bad to select them. For SZ1, select v'1 and f'1.
Press the delete key, or use the popup menu Channels > Mark selected as bad.
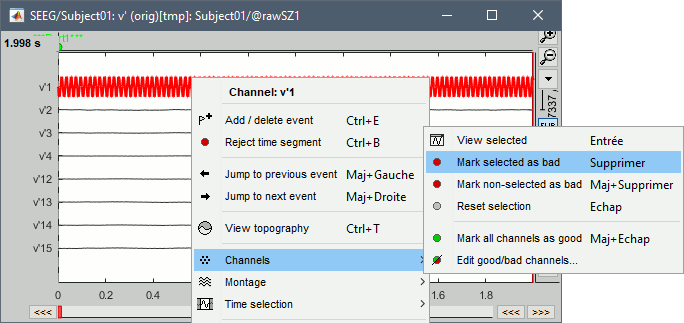
- Close the figure and repeat for the two other files.
- Bad channels for the three seizures:
SZ1: v'1, f'1
SZ2: v'1
SZ3: o'1
Edit event markers
Seizure onset
Two events are already available in this file: "Part1" only indicates the beginning of the file, and "Seizure" indicates approximately the onset of the first seizure. This marking was done for clinical use, to jump quickly to the page of interest, but here we need a more accurate marker. We will create a new marker "Onset" where we observe the beginning of the fast gamma activity, which is the main feature used computation of the epileptogenicity maps is based on.
Double-click on SZ1/Link to raw file to open the SEEG recordings for the first seizure, or do this immediately after marking the bad channels.
Switch to a bipolar montage: bipolar-1 is sparse (eg. g2-g1, g4-3), bipolar-2 is exhaustive (eg. g2-g1, g3-g2, g4-3), depending on how dense you want your figure to be.
In the Record tab, click on the event group "Seizure", then on the event timing. This will jump quickly to the page on which we will place our Onset marker.
Zoom around the g' contacts (using the buttons on the right of the figure or the corresponding shortcuts, indicated if you leave your mouse over the buttons for a second), or select the g' electrode in the montage menu.
- If needed, zoom locally in time in the current page (mouse wheel) and change the amplitude scale (right-click+mouse up/down).
Create an event group "Onset": Record tab > Events > Add group > "Onset", or select it.
Place the time cursor at 120.800s, add an Onset marker at this time (press the key "E", or use the popup menu Add/delete event).
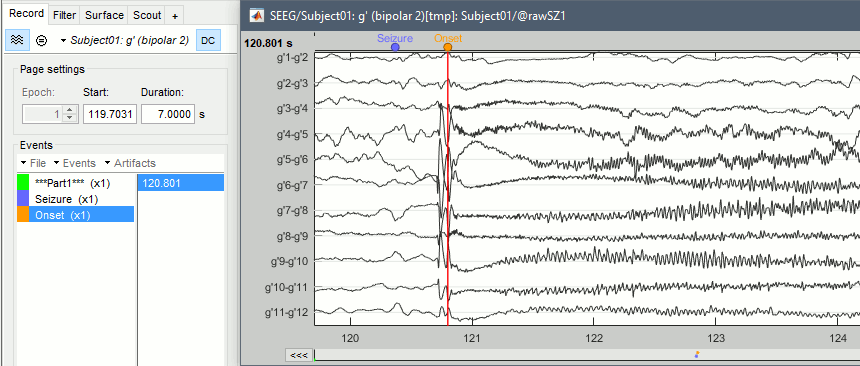
If you are marking a lot of events, you might be interest in using custom shortcuts.
Baseline
In the following sections, we will compute a time-frequency decomposition of the seizure and normalize it with respect to a baseline level. We will identify and import this baseline now. The baseline could be a segment of 5s to 30s of recordings, typically before the seizure, that contains as little interictal epileptiform activity as possible.
The duration of the baseline influences the computation time for the following steps of this tutorial. We will work here with short segments of 5s for illustration purposes, between 72.8s and 77.8s. Use longer segments for more accurate results.
.
.
.
Import epochs of interest
Time-frequency analysis
Epileptogenicity maps
Volume
Surface
Guidelines panel
Easy way for reproducing all these steps
On the hard drive
IntraElectrodes structure
Volume coregistration
Editing the contacts positions
Creating a new implantation file
Same as previous section, but not starting from an existing of contacts.
Importing realistic surfaces
TODO
- Fix all the links in this page
- Proofread and check spelling