|
Size: 2695
Comment:
|
Size: 4179
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Tutorial XX: Synchronization with Eyetracker data (event transfer) = ''Authors: Francois Tadel, Martin Völker'' |
= Synchronization with eyetracker (event transfer) = ''Authors: Martin Völker, Francois Tadel'' |
| Line 4: | Line 4: |
| This tutorial introduces how to transfer events from one data set to a second data set of a different type, which was recorded at the same time. We will explain this on the example of synchronizing ocular data recorded with an '''EyeLink '''eye tracker (''SR Research, Canada'') with a second raw recording, e.g. EEG. |
This tutorial introduces how to transfer events from one data set to a second data set of a different type, which was recorded at the same time but with a different sampling rate. We will explain this on the example of synchronizing ocular data recorded with an '''EyeLink '''eye tracker (''SR Research, Canada'') with a second raw recording, e.g. EEG. |
| Line 11: | Line 9: |
| Eye tracking is useful to detect blinks, saccades, microsaccades and fixations during an experiment. Typically, the eye tracker software will itself mark the most common artifact typeslike blinks and saccades. | Eye tracking is useful to detect blinks, saccades, microsaccades and fixations during an experiment. Typically, the eye tracker software will itself mark the most common artifact types like blinks and saccades. |
| Line 13: | Line 11: |
| These artifact markers are very useful for the analysis of EEG / MEG data, since specific time periods which are contominated with artifacts can be marked and excluded easily. To do so, we need to synchronize the eye tracker data with our EEG / MEG recordings. | These artifact markers are very useful for the analysis of EEG / MEG data, since specific time periods which are contaminated with artifacts can be marked and excluded easily. To do so, we need to synchronize the eye tracker data with our EEG / MEG recordings. |
| Line 15: | Line 13: |
| This tutorial focusses on how to snychronize EyeLink data (.edf) within brainstorm with raw data within brainstorm. Information about the '''EyeLink '''can be found at ''http://www.sr-research.com/index.html.'' |
This tutorial focusses on how to snychronize EyeLink data (.edf) with any other raw data type within brainstorm. Information about the '''EyeLink '''can be found at ''http://www.sr-research.com/index.html.'' |
| Line 20: | Line 16: |
| For this function to work, you need specific markers, which are stored within '''both''' EEG/MEG '''and''' eye tracker data '''at the same time.''' The exactly same number of this marker type has to be available in both data sets. They can, however, have different names. | For this function to work, you need specific markers, which are stored within '''both''' EEG/MEG '''and''' eye tracker data '''at the same time.''' The exact same number of this marker type has to be available in both data sets. They can, however, have different names. |
| Line 23: | Line 19: |
| First of all, the raw recording has to be linked to your brainstorm database, if not already done ([[Review continuous recordings]]). | First of all, the continuous MEG/EEG recording has to be linked to your Brainstorm database, if not already done ([[Tutorials/ReviewRaw|Review continuous recordings]]). |
| Line 25: | Line 21: |
| The EyeLink .edf file can be linked the same as the EEG recordings: | Then link the EyeLink .edf file in the same subject: |
| Line 27: | Line 23: |
| * Switch to the "functional data" view. <<BR>><<BR>> | * Switch to the "functional data" view. |
| Line 29: | Line 26: |
| * Select the file format: "'''EyeLink eye tracker (*.edf)'''" * Select the .edf file(s) * Click "open" * Due to the file format, the loading can take a while |
* Select the file format: "'''EyeLink eye tracker (*.edf)'''" * Select the EyeLink .edf file(s) * Click [open]. Due to the file format, the loading process can take a while. |
| Line 37: | Line 36: |
| * Into "Files A", drag and drop the raw data files from which you want to copy the events (in this case, the EyeLink files) * Into "Files B", drag and drop the recordings into which you want to paste the events; here, the EEG recordings. |
* In FilesA: drag and drop the files from which you want to copy the events (the EyeLink files). |
| Line 40: | Line 38: |
| [[attachment:evttransferTut_p1.png]] | * In FilesB: drag and drop the recordings into which you want to paste the events (the EEG files).<<BR>><<BR>> {{attachment:evttransferTut_p1.png||height="314",width="535"}} |
| Line 42: | Line 40: |
| It does not matter if the number of files in A and B is different. However, the number of sync markers within each data set has to match. | * It does not matter if the number of files in A and B is different. However, the total number of sync markers as to match exactly (number of markers in A = total number of markers in B). |
| Line 44: | Line 42: |
| == Process options == ... |
* Click on [RUN]. Select process "'''Synchronize > Transfer events (from A to B)'''" * In this example, the synchronization event is called "'''5'''" in the EyeLink file and "'''E5'''" in the EEG file. <<BR>><<BR>> {{attachment:evttransferTut_p2.png||height="294",width="352"}} * Click on [Run]. The function will calculate the offsets between the datasets with the help of the Sync events, which are known to be occuring at the exact same time. * In the Matlab command window, the variance and standard deviations of the sample offset is displayed per file of the destination data set.<<BR>><<BR>> {{attachment:evttransferTut_p3.png||height="141",width="529"}} * If the variance is too high, something is wrong with the sync events. For example, when the files in "B" are in the wrong order, the offsets will differ a lot and thus have a high variance. Depending on the timing of your recording and marker system, small variations are however perfectly normal. == Verification == It is important to check if the event transfer was executed properly: * If your recordings contain EOG channels, you can e.g. check if the blinks are marked correctly. <<BR>><<BR>> {{attachment:evttransferTut_p4.png||height="371",width="455"}} * If the two data sets share another event (besides the sync event you specified before), you can check if those are always marked at the same times. * Control if the number of events in each file meets your expectations. |
| Line 48: | Line 61: |
| * Forum: Development of eye tracker support http://neuroimage.usc.edu/forums/showthread.php?2193-Is-eye-tracker-data-support-interesting-to-Brainstorm | * Forum: [[http://neuroimage.usc.edu/forums/showthread.php?2193|Development of the eye tracker support]] |
Synchronization with eyetracker (event transfer)
Authors: Martin Völker, Francois Tadel
This tutorial introduces how to transfer events from one data set to a second data set of a different type, which was recorded at the same time but with a different sampling rate. We will explain this on the example of synchronizing ocular data recorded with an EyeLink eye tracker (SR Research, Canada) with a second raw recording, e.g. EEG.
Introduction
Eye tracking is useful to detect blinks, saccades, microsaccades and fixations during an experiment. Typically, the eye tracker software will itself mark the most common artifact types like blinks and saccades.
These artifact markers are very useful for the analysis of EEG / MEG data, since specific time periods which are contaminated with artifacts can be marked and excluded easily. To do so, we need to synchronize the eye tracker data with our EEG / MEG recordings.
This tutorial focusses on how to snychronize EyeLink data (.edf) with any other raw data type within brainstorm. Information about the EyeLink can be found at http://www.sr-research.com/index.html.
Requirements
For this function to work, you need specific markers, which are stored within both EEG/MEG and eye tracker data at the same time. The exact same number of this marker type has to be available in both data sets. They can, however, have different names.
Data import
First of all, the continuous MEG/EEG recording has to be linked to your Brainstorm database, if not already done (Review continuous recordings).
Then link the EyeLink .edf file in the same subject:
- Switch to the "functional data" view.
Right-click on the subject folder > Review raw file
Select the file format: "EyeLink eye tracker (*.edf)"
Select the EyeLink .edf file(s)
- Click [open]. Due to the file format, the loading process can take a while.
Event transfer
Now click on the "Process2" tab.
In FilesA: drag and drop the files from which you want to copy the events (the EyeLink files).
In FilesB: drag and drop the recordings into which you want to paste the events (the EEG files).
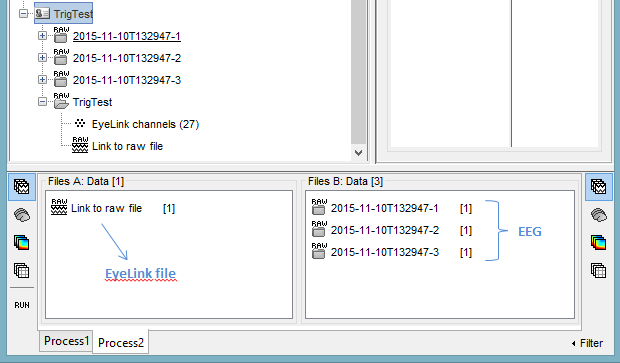
- It does not matter if the number of files in A and B is different. However, the total number of sync markers as to match exactly (number of markers in A = total number of markers in B).
Click on [RUN]. Select process "Synchronize > Transfer events (from A to B)"
In this example, the synchronization event is called "5" in the EyeLink file and "E5" in the EEG file.
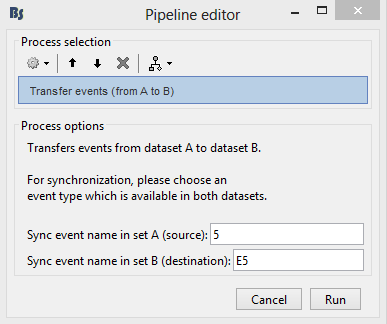
- Click on [Run]. The function will calculate the offsets between the datasets with the help of the Sync events, which are known to be occuring at the exact same time.
In the Matlab command window, the variance and standard deviations of the sample offset is displayed per file of the destination data set.
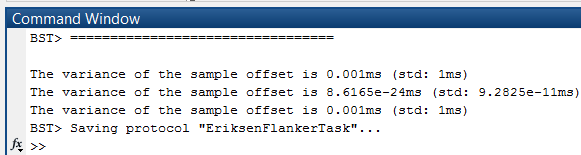
- If the variance is too high, something is wrong with the sync events. For example, when the files in "B" are in the wrong order, the offsets will differ a lot and thus have a high variance. Depending on the timing of your recording and marker system, small variations are however perfectly normal.
Verification
It is important to check if the event transfer was executed properly:
If your recordings contain EOG channels, you can e.g. check if the blinks are marked correctly.
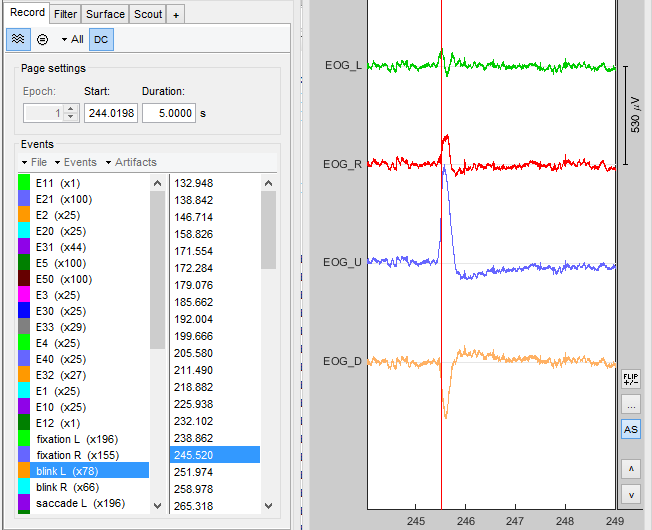
- If the two data sets share another event (besides the sync event you specified before), you can check if those are always marked at the same times.
- Control if the number of events in each file meets your expectations.
