|
Size: 2352
Comment:
|
Size: 5772
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Use FreeSurfer cortical parcellation = The free software !FreeSurfer can be used to extract brain and head surfaces from a T1 MRI. It also automatically registers the individual cortex surfaces to two atlases (Desikan-Killiany and Destrieux). The results of those of those cortex segmentations can now be imported in Brainstorm as scouts, and therefore be used as anatomical ROIs. |
= Using FreeSurfer = The free software !FreeSurfer can be used to extract brain and head surfaces from a T1 MRI. It also automatically registers the individual cortex surfaces to two atlases (Desikan-Killiany and Destrieux). The process is fully automatic and the results can be imported in Brainstorm with just a few mouse clicks. |
| Line 4: | Line 4: |
| For more information about the methods involved and the software installation: | == Running FreeSurfer == 1. Downloading and installing FreeSurfer is very easy. It just takes some time because the distribution package is huge (get ready to download several Gb).<<BR>>Just follow the instructions: http://surfer.nmr.mgh.harvard.edu/fswiki/DownloadAndInstall 1. Set up the FreeSurfer environment: * setenv FREESURFER_HOME /.../local/freesurfer * setenv SUBJECTS_DIR /.../data/freesurfer/subjects * setenv FUNCTIONALS_DIR /.../data/freesurfer/sessions * cd /.../local/freesurfer * source FreeSurferEnv.csh |
| Line 6: | Line 13: |
| * Cortical parcellation: http://freesurfer.net/fswiki/CorticalParcellation * !FreeSurfer Wiki: http://surfer.nmr.mgh.harvard.edu/fswiki/ |
1. Run the reconstruction: * recon-all -i <mri_file> -subjid <subject_id> * recon-all -all -subjid <subject_id> 1. Leave your office, come back the next day. The process is fully automatic, but quite resource consuming. 1. Done. Everything is ready to be imported in Brainstorm. The results are usually perfect, but depending on the quality of the structural MR, it may fail. Because of this unpredictable behavior, you always need to check visually the final surfaces. The FreeSurfer wiki suggests that you check all the steps with FreeSurfer. We suggest instead that you load it all in Brainstorm and go back to the manual checking/editing only if it looks bad. 1. More detailed instructions for setting up the environment and tuning the reconstruction here: http://surfer.nmr.mgh.harvard.edu/fswiki/RecommendedReconstruction<<BR>>More questions about FreeSurfer: Use the text box on the top right corner of the previous page to search the wiki. |
| Line 9: | Line 20: |
| == Import parcellations == | == Cortical parcellations == Cortical parcellation: http://freesurfer.net/fswiki/CorticalParcellation |
| Line 16: | Line 31: |
| * Right hemisphere: /surf/lh.pial (or lh.smoothwm) 1. Add the atlas-based parcellations to each hemisphere <<BR>><<BR>> |
* Right hemisphere: /surf/lh.pial (or lh.smoothwm)<<BR>><<BR>> 1. Add the atlas-based parcellations to each hemisphere <<BR>><<BR>> {{attachment:importAnnot1.gif}} |
| Line 23: | Line 38: |
| * The .annot files are read with the !FreeSurfer read_annotation.m Matlab function, then converted to Brainstorm scouts, and saved in the surface files in the "Scout" field | * The .annot files are read with the !FreeSurfer read_annotation.m Matlab function, then converted to Brainstorm scouts, and saved in the surface files in the "Scout" field <<BR>><<BR>> |
| Line 27: | Line 42: |
| 1. Double-click on "cortex", go to the Scout tab, and select a few scouts, to check that the .annot files where imported properly | 1. Double-click on "cortex", go to the Scout tab, and select a few scouts, to check that the .annot files where imported properly<<BR>><<BR>> {{attachment:importAnnot2.gif}} |
| Line 32: | Line 47: |
| {{attachment:dk_fs.jpg}} |
|
| Line 33: | Line 50: |
{{attachment:dk_bs.gif}} |
|
| Line 36: | Line 55: |
| Destrieux atlas, in !FreeSurfer | {{attachment:dk_bs2.gif}} == Parcellations for the default anatomy (MNI/Colin27) == The !FreeSurfer cortex and white matter surfaces are available in the standard distribution of Brainstorm, they come with the all the !FreeSurfer parcellations. You can use those surfaces instead of the default Brainstorm ones, which are created with BrainVISA, which do not include the Desikan-Killiany and Destrieux atlases. 1. In any protocol, go to the anatomy view, and select the "Default anatomy" folder.<<BR>><<BR>> {{attachment:defaultAnat.gif}} 1. If you do not see the surfaces "!FreeSurfer: cortex *" and "!FreeSurfer: white *", your protocol is not using the most recent version of the Brainstorm default anatomy. To update: * Make sure you are running the latest version of Brainstorm * Right-click on "Default anatomy" > Use default > MNI_Colin27 * Close the MRI Viewer when it opens. Now you should see all the !FreeSurfer surfaces 1. Make one of the !FreeSurfer surfaces as your default cortex surface (for instance, the downsampled grey/csf interface): * Right-click on "!FreeSurfer: cortex 15000V" > Set surface type > Cortex * Right-click on the top "cortex" file > Set surface type > Other (this is not necessary, but it avoids ambiguity) * To make sure that everything is updated properly: Right-click on "Default anatomy" > Reload<<BR>><<BR>> {{attachment:defaultAnat2.gif}} 1. Double-click on the new default cortex (!FreeSurfer: cortex 15000V) to open it 1. In the "Scout" tab, click on the "Load scout" button (on the right of the scouts list)<<BR>><<BR>> {{attachment:importScout.gif}} 1. In the list of available scout files, pick on that matches the surface that is currently displayed: '''scout_freesurfer_*_15000V.mat'''<<BR>><<BR>> {{attachment:figureCortex.gif}} 1. You can now run source analysis (computation forward and inverse models) using this new cortex surface, and use the !FreeSurfer atlases as regions of interest |
Using FreeSurfer
The free software FreeSurfer can be used to extract brain and head surfaces from a T1 MRI. It also automatically registers the individual cortex surfaces to two atlases (Desikan-Killiany and Destrieux). The process is fully automatic and the results can be imported in Brainstorm with just a few mouse clicks.
Running FreeSurfer
Downloading and installing FreeSurfer is very easy. It just takes some time because the distribution package is huge (get ready to download several Gb).
Just follow the instructions: http://surfer.nmr.mgh.harvard.edu/fswiki/DownloadAndInstallSet up the FreeSurfer environment:
- setenv FREESURFER_HOME /.../local/freesurfer
- setenv SUBJECTS_DIR /.../data/freesurfer/subjects
- setenv FUNCTIONALS_DIR /.../data/freesurfer/sessions
- cd /.../local/freesurfer
source FreeSurferEnv.csh
- Run the reconstruction:
recon-all -i <mri_file> -subjid <subject_id>
recon-all -all -subjid <subject_id>
- Leave your office, come back the next day. The process is fully automatic, but quite resource consuming.
Done. Everything is ready to be imported in Brainstorm. The results are usually perfect, but depending on the quality of the structural MR, it may fail. Because of this unpredictable behavior, you always need to check visually the final surfaces. The FreeSurfer wiki suggests that you check all the steps with FreeSurfer. We suggest instead that you load it all in Brainstorm and go back to the manual checking/editing only if it looks bad.
More detailed instructions for setting up the environment and tuning the reconstruction here: http://surfer.nmr.mgh.harvard.edu/fswiki/RecommendedReconstruction<<BR>>More questions about FreeSurfer: Use the text box on the top right corner of the previous page to search the wiki.
Cortical parcellations
Cortical parcellation: http://freesurfer.net/fswiki/CorticalParcellation
Process your subject MRI with the full FreeSurfer T1 analysis pipeline
- In Brainstorm, create a protocol and a subject, as explained in the introduction tutorials
Import the following files from the FreeSurfer architecture:
- MRI: /mri/T1.mgz
- Head surface: /bem/wateshed/outer_skin_surface
- Left hemisphere: /surf/lh.pial (or lh.smoothwm)
Right hemisphere: /surf/lh.pial (or lh.smoothwm)
Add the atlas-based parcellations to each hemisphere
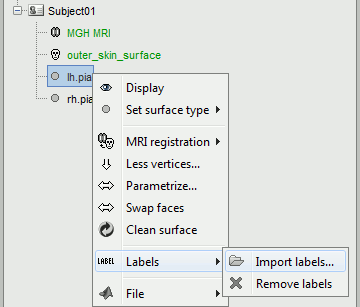
Right-click on "lh" > Labels > Import labels
- Select one of the annotation files in the /label/ folder:
- Destrieux atlas: lh.aparc.a2009s.annot
- Desikan-Killiany atlas: lh.aparc.annot
- Repeat the same operation for the right hemisphere
The .annot files are read with the FreeSurfer read_annotation.m Matlab function, then converted to Brainstorm scouts, and saved in the surface files in the "Scout" field
Downsample each hemisphere to 7500 vertices (the scouts will be downsampled accordingly)
Right-click > Less verticesConcatenate the two downsampled surfaces
Select lh_7500 and rh_7500 > right-click > Merge surfaces- Rename the new surface into "cortex". This last surface contains all the scouts (ie. ROIs) defined for lh and rh, which will be loaded automatically when you display the surface in Brainstorm.
Double-click on "cortex", go to the Scout tab, and select a few scouts, to check that the .annot files where imported properly
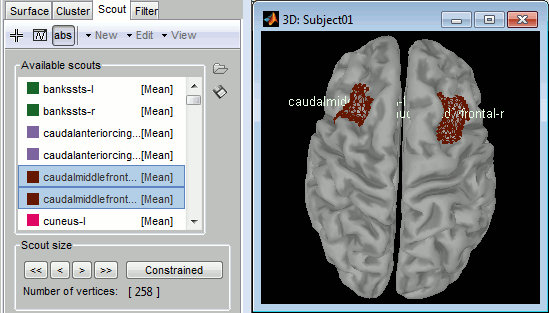
Examples
Desikan-Killiany atlas, in FreeSurfer:

Desikan-Killiany atlas, in Brainstorm:
Desikan-Killiany atlas, in Brainstorm after downsampling to 15000 vertices:
Parcellations for the default anatomy (MNI/Colin27)
The FreeSurfer cortex and white matter surfaces are available in the standard distribution of Brainstorm, they come with the all the FreeSurfer parcellations. You can use those surfaces instead of the default Brainstorm ones, which are created with BrainVISA, which do not include the Desikan-Killiany and Destrieux atlases.
In any protocol, go to the anatomy view, and select the "Default anatomy" folder.
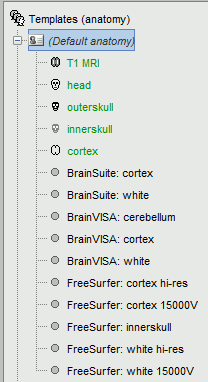
If you do not see the surfaces "FreeSurfer: cortex *" and "FreeSurfer: white *", your protocol is not using the most recent version of the Brainstorm default anatomy. To update:
- Make sure you are running the latest version of Brainstorm
Right-click on "Default anatomy" > Use default > MNI_Colin27
Close the MRI Viewer when it opens. Now you should see all the FreeSurfer surfaces
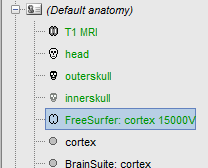
Make one of the FreeSurfer surfaces as your default cortex surface (for instance, the downsampled grey/csf interface):
Right-click on "FreeSurfer: cortex 15000V" > Set surface type > Cortex
Right-click on the top "cortex" file > Set surface type > Other (this is not necessary, but it avoids ambiguity)
To make sure that everything is updated properly: Right-click on "Default anatomy" > Reload
Double-click on the new default cortex (FreeSurfer: cortex 15000V) to open it
In the "Scout" tab, click on the "Load scout" button (on the right of the scouts list)
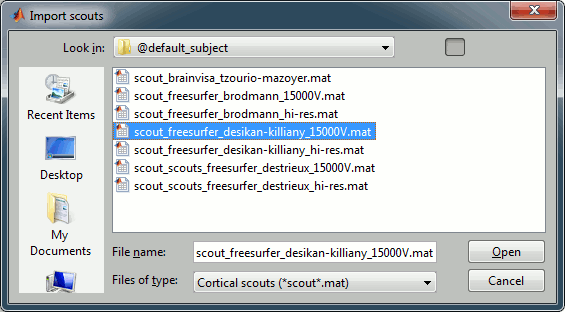
In the list of available scout files, pick on that matches the surface that is currently displayed: scout_freesurfer_*_15000V.mat
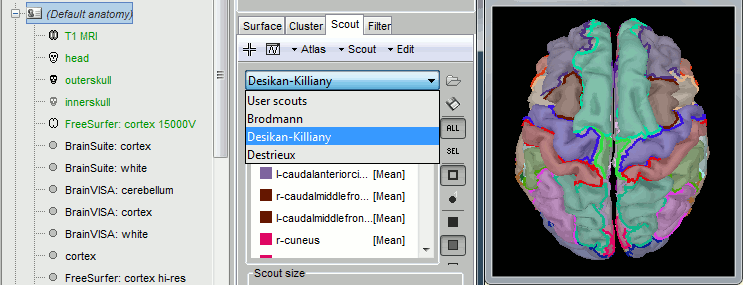
You can now run source analysis (computation forward and inverse models) using this new cortex surface, and use the FreeSurfer atlases as regions of interest
