|
Size: 2539
Comment:
|
Size: 2503
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 16: | Line 16: |
| * Right hemisphere: /surf/lh.pial (or lh.smoothwm) 1. Add the atlas-based parcellations to each hemisphere <<BR>><<BR>>{{attachment:importAnnot1.gif}} |
* Right hemisphere: /surf/lh.pial (or lh.smoothwm)<<BR>><<BR>> 1. Add the atlas-based parcellations to each hemisphere <<BR>><<BR>> {{attachment:importAnnot1.gif}} |
| Line 23: | Line 23: |
| * The .annot files are read with the !FreeSurfer read_annotation.m Matlab function, then converted to Brainstorm scouts, and saved in the surface files in the "Scout" field | * The .annot files are read with the !FreeSurfer read_annotation.m Matlab function, then converted to Brainstorm scouts, and saved in the surface files in the "Scout" field <<BR>><<BR>> |
| Line 27: | Line 27: |
| 1. Double-click on "cortex", go to the Scout tab, and select a few scouts, to check that the .annot files where imported properly<<BR>><<BR>>{{attachment:importAnnot2.gif}} | 1. Double-click on "cortex", go to the Scout tab, and select a few scouts, to check that the .annot files where imported properly<<BR>><<BR>> {{attachment:importAnnot2.gif}} |
| Line 36: | Line 36: |
| {{attachment:dk_bs.jpg}} | {{attachment:dk_bs.gif}} |
| Line 41: | Line 41: |
Destrieux atlas, in !FreeSurfer {{attachment:de_fs.gif}} |
Use FreeSurfer cortical parcellation
The free software FreeSurfer can be used to extract brain and head surfaces from a T1 MRI. It also automatically registers the individual cortex surfaces to two atlases (Desikan-Killiany and Destrieux). The results of those of those cortex segmentations can now be imported in Brainstorm as scouts, and therefore be used as anatomical ROIs.
For more information about the methods involved and the software installation:
Cortical parcellation: http://freesurfer.net/fswiki/CorticalParcellation
FreeSurfer Wiki: http://surfer.nmr.mgh.harvard.edu/fswiki/
Import parcellations
Process your subject MRI with the full FreeSurfer T1 analysis pipeline
- In Brainstorm, create a protocol and a subject, as explained in the introduction tutorials
Import the following files from the FreeSurfer architecture:
- MRI: /mri/T1.mgz
- Head surface: /bem/wateshed/outer_skin_surface
- Left hemisphere: /surf/lh.pial (or lh.smoothwm)
Right hemisphere: /surf/lh.pial (or lh.smoothwm)
Add the atlas-based parcellations to each hemisphere
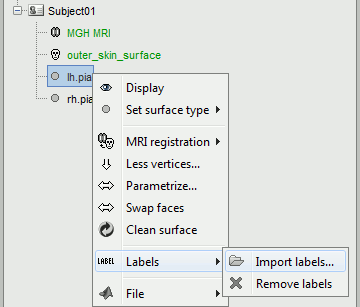
Right-click on "lh" > Labels > Import labels
- Select one of the annotation files in the /label/ folder:
- Destrieux atlas: lh.aparc.a2009s.annot
- Desikan-Killiany atlas: lh.aparc.annot
- Repeat the same operation for the right hemisphere
The .annot files are read with the FreeSurfer read_annotation.m Matlab function, then converted to Brainstorm scouts, and saved in the surface files in the "Scout" field
Downsample each hemisphere to 7500 vertices (the scouts will be downsampled accordingly)
Right-click > Less verticesConcatenate the two downsampled surfaces
Select lh_7500 and rh_7500 > right-click > Merge surfaces- Rename the new surface into "cortex". This last surface contains all the scouts (ie. ROIs) defined for lh and rh, which will be loaded automatically when you display the surface in Brainstorm.
Double-click on "cortex", go to the Scout tab, and select a few scouts, to check that the .annot files where imported properly
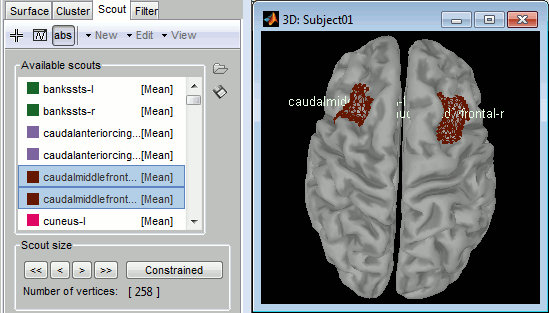
Examples
Desikan-Killiany atlas, in FreeSurfer:

Desikan-Killiany atlas, in Brainstorm:
Desikan-Killiany atlas, in Brainstorm after downsampling to 15000 vertices:
