|
Size: 88336
Comment:
|
Size: 19792
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 10: | Line 10: |
| == License == This tutorial dataset (MEG and MRI data) remains a property of the MEG Lab, McConnell Brain Imaging Center, Montreal Neurological Institute, McGill University, Canada. Its use and transfer outside the Brainstorm tutorial, e.g. for research purposes, is prohibited without written consent from the MEG Lab. If you reference this dataset in your publications, please aknowledge its authors (Elizabeth Bock, Esther Florin, Francois Tadel and Sylvain Baillet) and cite Brainstorm as indicated on the [[http://neuroimage.usc.edu/brainstorm/CiteBrainstorm|website]]. For questions, please contact us through the forum. |
|
| Line 18: | Line 13: |
| * Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder). This is really important that you always keep your original data files in a separate folder: the program folder can be deleted when updating the software, and the contents of the database folder is supposed to be manipulated only by the program itself. * Start Brainstorm (Matlab scripts or stand-alone version) |
* Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder). * Start Brainstorm (Matlab scripts or stand-alone version). |
| Line 21: | Line 16: |
| * "'''No, use individual anatomy'''", * "'''Yes, use one channel file per subject'''". |
* '''No, use individual anatomy''', * '''No, use one channel file per run'''. |
| Line 25: | Line 20: |
| * Create a new subject Subject01 * In the anatomy view, right-click on the subject > Import anatomy folder, select the file format "FreeSurfer folder" and select the folder "sample_raw/Anatomy". This imports all the available files in a segmentation folder generated with FreeSurfer, as illustrated in [[http://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer|this page]]. * Leave the default number of vertices of the cortex surface (15000) * When the MRI Viewer window shows up, you have to define the position of the 6 fiducial points, as described in CoordinateSystems (set the SCS fiducials as the read anatomical nasion/left/right, not the position of the CTF coils). At the end, you should have the following MRI coordinates (+/- a few millimeters): |
* Create a new subject Subject01. * Right-click on the subject node > Import anatomy folder: * Set the file format: "FreeSurfer folder" * Select the folder: '''sample_raw/anatomy''' * Number of vertices of the cortex surface: 15000 (default value) * Click on the link "'''Click here to compute MNI transformation'''". * Set the 3 required fiducial points (indicated in MRI coordinates): |
| Line 32: | Line 31: |
| * AC: x=129, y=137, z=157 * PC: x=129, y=113, z=157 * IH: x=129, y=118, z=209 (anywhere on the midsagittal plane) |
|
| Line 36: | Line 32: |
| * Click on Save when you're done, and wait until the process is over. At the end, a figure shows the surfaces that were generated. Close it. . {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=anat.gif|anat.gif|class="attachment"}} * Four files are now available as the anatomy of Subject01: a T1 MRI, a head surface, and two cortex surfaces (high and low resolution). The anatomy of the subject is ready. |
* At the end of the process, make sure that the file "cortex_15000V" is selected (downsampled pial surface, that will be used for the source estimation). If it is not, double-click on it to select it as the default cortex surface. <<BR>><<BR>> {{attachment:anat.gif||height="175",width="368"}} |
| Line 40: | Line 34: |
| ==== Without the individual MRI ==== If you do not have access to an individual MR scan of the subject (or if its quality is too low to be processed with FreeSurfer), but if you have digitized the head shape of the subject using a tracking system, you have an alternative: deform one of the Brainstorm templates (Colin27 or ICBM152) to match the shape of the subject's head. |
== Link the recordings == * Switch to the "functional data" view (middle button in the toolbar above the database explorer). * Right click on the Subject01 > '''Review raw file''': * Select the file format: '''MEG/EEG: CTF''' * Select the folder: '''sample_raw/Data/subj001_somatosensory_20111109_01_AUX-f.ds''' * Refine the registration with the head points: '''YES'''.<<BR>><<BR>> {{attachment:refineBefore.gif||height="229",width="200"}} {{attachment:refineAfter.gif||height="229",width="200"}} |
| Line 43: | Line 41: |
| . For more information, read the following tutorial: [[http://neuroimage.usc.edu/brainstorm/Tutorials/TutWarping|Warping default anatomy]] == Access the raw file == The basic tutorials you read before explain how to import recordings in the database: this operation creates a copy of all the data in Matlab .mat files in the Brainstorm database folders. You could process continuous recordings in the same way, but the .mat format has this limitation that the entire file has to be read even when you want to access just a portion of it. Long recordings usually cannot fit in memory and have to be split in small blocks of a few seconds, which makes it very difficult to review and process. Brainstorm offers the possibility to visualize continuous MEG/EEG recordings in any of the supported file formats without having to fully "import" them. A link to the native file is created in the database, which can be then manipulated almost like the "imported" recording blocks. Only the description of the file is saved in the database, and when displaying it the values are read directly from the native file. In addition, an interface allows to edit the time markers that are saved in the file. These markers can then be used to import the recordings in the database (ie. to do the segmentation of the continuous recordings in epochs/trials). Then the imported epochs/trials (hard copies in .mat format) can be pre-processed and averaged. * Select the "Functional data (sorted by subject)" exploration mode (second button in the toolbar above the database explorer). * Right-click on the subject node, and select: "Review raw file". Select the "MEG: CTF" file type, and pick the ds folder in "/sample_raw/Data". . {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=menuReview.gif|menuReview.gif|class="attachment"}} * Then you're asked if you want to "Refine the registration with the head points". This operation improves the initial MRI/MEG registration by fitting the head points digitized before the MEG acquisition on the scalp surface with an ICP algorithm. Answer yes. Even if the result is not perfect, it usually improves the positioning of the head in the MEG helmet. The grey surface represents the head extracted from the MRI, the yellow surface represents the inside of the MEG helmet, and the green dots are the head shape points digitized with the Polhemus device; the goal is to align the green points on the grey surface. . {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=refine.gif|refine.gif|class="attachment"}} {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=refineBefore.gif|refineBefore.gif|class="attachment"}} {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=refineAfter.gif|refineAfter.gif|class="attachment"}} * Two new files appeared in the database explorer: . {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=linkInTree.gif|linkInTree.gif|class="attachment"}} * The channel file contains the definition of the sensors, exactly as when importing the files in the database with the "Import MEG/EEG" menu. It is saved in the folder ''(Common files)'', because the subject was created using the option "Yes, use one channel file per subject". Therefore, the same channel file will be used for all the folders of Subject01. * The node named "Link to raw file" contains all the information that was read from the continuous file (file format, time vector, sampling frequency, events, bad channels, path to the original file, etc.), but no recordings. The MEG and EEG values recorded will be read directly from the native file. == Review the recordings == === Open the file === Right-click on the data file > MEG (all) > Display time series. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=displayTsMenu.gif|displayTsMenu.gif|class="attachment"}} You can see new information in the tab "Record" and a figure showing the recordings. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=rawPanel.gif|rawPanel.gif|class="attachment"}} === Navigate in time === As described in the basic tutorials, you can set the current time by using either the time panel (buttons and text field), or the figure (click on the white or grey areas of the figure). But you can notice that only a few seconds are visible in the figure, while the time panel (top left of the previous figure), indicates that we have 360s of recordings. Only a small block of the continuous file has been loaded in memory. This small time window can be configured with the tab '''Record/Page settings''', with the text boxes '''Start''' and '''Duration'''. The time series figure is similar to the ones that were presented in the previous tutorials, with a few new elements. The navigation bar at the bottom represents the time of the entire raw file, where the events are also represented by dots. The ''''<<<'''' and ''''>>>'''' buttons are the same as the ones in the time panel, and jump to the previous/next segment in the file. Clicking on the bar or dragging the red cursor change the current time window as well. === Sensor selection === Let's switch to a nicer representation of the recordings time series: click on the "Display mode" button in the toolbar of the Record tab. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=tsColumn.gif|tsColumn.gif|class="attachment"}} Now the traces are displayed in columns, but all the channels are displayed in the same figure, which makes it unreadable. Select a subset of channels by right-clicking on the figure > '''Montages''', with the drop-down menu in the Record tab or with a keyboard shortcut ('''Shift+A, B, C'''...). Default groups of sensors are available for some MEG systems, but you can also create your own groups of sensors with the menu "Edit montages". {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=tsChannelSelection.gif|tsChannelSelection.gif|class="attachment"}} === Amplitude scale === In this display mode, the amplitude scale is represented on the right of the figure. You can adjust this vertical scale: * Use the buttons "'''^'''" and "'''v'''" on the right side of the figure. The shortcuts for these buttons are indicated in the tooltips (leave the mouse for a short while over a button) * Hold the '''Shift key''' and move the mouse wheel, or use the keys "'''+'''" and "'''-'''". * Use the button "'''...'''" on the right side of the figure ("Set scale manually") to set the scale to a precise level. When scrolling in time to a different page, the amplitude scale is by default kept. You can change this behavior to automatically re-evaluate an optimal scale each time you change the current time window. This option is called "Auto-scale amplitude" and is disabled by default. To activate it: click on the "'''AS'''" button on the right of the figure, or check the menu "'''Display > Auto-scale amplitude'''" in the Record tab. === Display options === . {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=displayOptions.gif|displayOptions.gif|class="attachment"}} * '''Remove DC offset''': Button [DC] in the Record tab. When selected, for each channel, the average value over the entire current time window is subtracted from the channel values. This means that if you change the length of the time window, the value that is removed from each channel may change. It doesn't make much sense to disable this option for unprocessed MEG recordings. * '''Apply CTF compensation''': Button [CTF] in the Record tab. Enable/disable the CTF noise correction based on the reference sensors, when it is not already applied in the file. In the current file, the CTF 3rd order gradient compensation is already applied, therefore this option is not available. * '''Flip +/-''': Button in the right part of the time series figure. Exchange the direction of the Y axis, useful mostly for clinical EEG. * '''Set scale manually''': Button [...] in the figure. Forces a defined amplitude scaling. * '''Auto-scale amplitude''': Button [AS] in the figure. When selected, the vertical scale is adapted to the maximum value over the time window when the time window changes. When not selected: the vertical scales keeps its last value when you jump to another part of the file. === Filters for visualization === With the Filter tab, you can apply a band-pass filter to the recordings, or remove a set of specific frequencies (example: the 50Hz or 60Hz power lines contamination and their harmonics). The filters are applied only to the time window that is currently loaded; hence if the segment is too short for the required filters, the results could be inaccurate. The option "'''Mirror signal before filtering'''" triples artificially the length of the signal with a mirror symmetry on each side, to avoid the strong edge effects that these filters can generate. These visualization filters are not very accurate, they just provide a quick estimate for visualization only, the results are not saved anywhere. To filter properly the continuous files, please use the Process1 tab. After testing the high-pass, low-pass and notch filters, uncheck them. If not you will probably forget about them, and they will stay on until you restart Brainstorm. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=onlineFilter.gif|onlineFilter.gif|class="attachment"}} == Events, markers, triggers == === Lists of events === You probably noticed little green and blue dots on top of the recordings in the MEG figure. They represent the triggers of the electric stimulation. The stimulation computer sent these triggers simultaneously to the electric stimulator, which converted them into electric pulses sent to the wrists of the subject, and to the acquisition computer, which recorded them together with the values of the MEG sensors. All the temporal markers that are available in the file are listed in the Recordings section of the Record tab: * on the left, the groups of events and the number of occurrence for each group (102 stimulations on the left, 98 on the right); * on the right, all the occurrences of the selected event group, described by the time instant at which they occur. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=eventSelect.gif|eventSelect.gif|class="attachment"}} These two lists are interactive. If you click on a event group (left list), it shows the occurrences for the selected event group in the right list. If you click on one particular event in the right list, the current time is set in the MEG figure to the selected event. If you click on a dot representing an event in the MEG figure, the corresponding event group and occurrence are selected in the Record tab. These "events" can represent either stimulation triggers that were recorded during the acquisition, or additional markers that were placed by the user during the analysis (eye blinks, epileptic spikes). === Adding events === First create a new group of events called "Test", with the menu "'''Events > Add group'''". It creates a new event group with no occurrences (x0). {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=addGroup.gif|addGroup.gif|class="attachment"}} Then select the event group "Test", and set the current time where you want to add a new Test even, by clicking on the figure (current time = where the vertical red line is). Add a few occurrences with any of the three methods: * In the Record tab: select the menu Events > '''Add / delete event ''' * In the time series figure: right-click > '''Add / delete event ''' * In the time series figure: press '''Ctrl + E ''' * Note that you can click outside of the white area to select the time (on top of the figure), or use the shortcut '''Shift+click'''. If the display is too dense, it can be difficult to set the current time instead of selecting a channel. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=addEvent_done.gif|addEvent_done.gif|class="attachment"}} Now remove all the events occurrences, but not the group "Test": * In the Record tab: select one or more event occurrences (hold Shift or Control + mouse click), and press the '''Delete''' '''key'''. * In the time series figure: click on the event to delete (on the blue dot), and right-click > '''Add / delete event ''' * In the time series figure: click on the event to delete, and press '''Ctrl + E''' === Extended events === You can also use this interface to create events that have a temporal extension, ie. they last for more than one time sample. This is usually used to define bad/artifacted segments in the recordings. * In the time series window, select a time range (click + move) instead of just setting the current time. * Note that you can click outside of the white area to select the time (on top of the figure). If the display is too dense, it can be difficult to select a time window instead of a channel. * Add an event (menus or Control+E): note the way it is represented in the figure and in the Event panel. * The first occurrence you add in an event group defines the event type: single time point, or time range. You cannot mix different types of events in a group. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=extEventSel.gif|extEventSel.gif|class="attachment"}} {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=extEvent.gif|extEvent.gif|class="attachment"}} === Custom shortcuts === When reviewing long recordings and adding manually lots of events (eg. when marking manually epileptic spikes), using the menus presented previously is not very convenient because they require many mouse clicks. Using the menu '''Events > Edit keyboard shortcuts''', you can associate custom events to the key 1 to 9 of the keyboard. Define the name of the event type to create for each key, and then simply press the corresponding key to add/delete a marker at the current time position. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=evtCustom.gif|evtCustom.gif|class="attachment"}} === Bad segments === It is very common to have portions of the recordings heavily contaminated by events coming from the subject (eye blinks, movements, heartbeats, teeth clenching...) or from the environment (stimulation equipment, elevators, cars, trains, building vibrations...). Some of them are well defined and can be removed efficiently, it is the purpose of the next tutorial, some cannot. For this last category, it is usually safer to mark the noisy segments as bad, and ignore them for the rest of the analysis. To mark a segment of recordings as bad, the procedure is the same as for defining an extended event: select a time window, and then tag it as bad with one of the following methods. * In the Record tab: select the menu '''Events > Reject time segment''', * In the time series figure: right-click > '''Reject time segment''', * In the time series figure: press '''Ctrl + B''' It creates a new event group BAD, and add an extended event to it. Later, when epoching this file (extracting time blocks around the markers and saving them in the database), the trials that contain a segment marked as bad will be imported but marked as bad, and ignored in the rest of the analysis. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=bad.gif|bad.gif|class="attachment"}} === Saving modifications === Now you can delete all the event groups that you've just created and leave only the "left" and "right" triggers: select the unwanted event groups and press the '''Delete key''', or use the menu '''Events > Delete group'''. When you close the raw file viewer, or the last figure that shows a part of the raw file, the dataset is unloaded, the file is released and the memory is freed. If you edited the events for this file, you are asked whether to save the modifications or not. If you answer "Yes", the modifications are saved only in the database link (Link to raw file), not in the original file itself. Therefore, you would see the changes the next time you double-click on the "link to raw file" again, but not if you open the original .ds file in another protocol or with an external program. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=saveModif.gif|saveModif.gif|class="attachment"}} Note that events you edit are not automatically saved until that moment. As you would do with any other type of computer work, save your work regularly, to limit the damages of a program or computer crash. In the Record tab, use the menu ''' File > Save modifications'''. === Other menus === {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=menuFile.gif|menuFile.gif|class="attachment"}} ==== File ==== * '''Import in database''': Import blocks of the current continuous file into the database. Equivalent to a right click on the "Link to raw file" in the database explorer > Import in database. * '''Save modifications''': Save the modifications made to the events in the database link (Link to raw file) * '''Add events from file''': Import events from an external file. Many file formats from various software are supported. * '''Read events from channel''': Read the information saved during the acquisition in an digital auxiliary channel (eg. a stimulus channel) and generate events. Generation of events based on an analog channel is not supported yet. * '''Export all events''': Save all the events in an external file, to use them in an external software or for another continuous file. * '''Export selected events''': Same as "Export all events" but exports only the selected event groups or event occurrences. ==== Events ==== * '''Rename group''': Rename the selected group of events. Shortcut: double-click. * '''Set color''': Change the color associated with an event group. * '''Merge groups''': Merge to event groups into a new group. Initial groups are deleted; to keep them, duplicate them before merging. * '''Combine stim/response''': Shortcut to call the process "Import recordings > Combine stim/response", to create new groups of events based on stim/response logic. Example: Stimulus A can be followed by response B or C. Use this process to split the group A in two groups: AB, followed by B; and AC, followed by C. * '''Duplicate groups''': Make a copy of the selected event groups. * '''Convert to simple events''': Convert a group of extended events (several time points for each event), to simple events (one time point). An option window asks you whether to keep the first or the last sample only of the extended events. * '''Edit keyboard shortcuts''': Custom associations between keys 1..9 and events * '''Reject time segment''': Mark the current time selection as bad (ie. create a new event type called BAD) * '''Jump to previous/next event''': Convenient way of browsing through all the occurrences of a event group. Shortcut: '''Shift + left/right''' == Shortcut summary == === Keyboard shortcuts === * '''Left / right arrows''': * No other key: Change current time, sample by sample * With '''Control '''key: Jump to previous/next time segment (same as the "<<<" and ">>>" buttons) * With '''Shift '''key: Jump to next event of the selected group * On MacOS, these shortcuts are different: please read the tooltips from the buttons ">", ">>", and ">>>" in the time panel to get the appropriate shortcuts. * '''Page-up / page-down''': * Same as left/right arrows, but faster (10 samples at a time) * If epochs are defined in the file: '''Control + page-up/page-down''' jumps to the next/previous epoch. * '''F3/Shift+F3''': Jump to the next/previous epoch or page * '''F4/Shift+F4''': Jump to the next/previous half-page * '''Plus / minus''': Adjust the vertical scale of the time series * '''Control + E''': Add / delete event occurrence * '''Control + T''': Open a 2D topography window at the current time * '''Shift + Letter''': Changes the set of electrodes currently displayed in the figure (list available by right-clicking on the figure > Display setup > ...) * '''Enter''': Display the selected channels in a separate figure (selected channels = lines on which you clicked, that are shown in red) * '''Escape''': Unselect all the selected channels * '''Delete''': Mark the selected channels as bad === Mouse shortcuts === * '''Mouse click on a channel''': Select the channel * '''Mouse click''': Change current time * '''Mouse click + Shift''': For the selection of the current time (do not select any sensor, even when clicking on a line) * '''Mouse click + move''': Select time range * '''Mouse wheel''': Zoom around current time * '''Control + mouse wheel''': Zoom vertically * '''Shift + mouse wheel''': Adjust the vertical scale of the time series * '''Right-click''': Display popup menu * '''Right-click + move''': Move in a zoomed figure * '''Double click''': Restore initial zoom settings (but do not restore the vertical scale of the time series) == Other views == === Topography views === Exactly as introduced in the [[http://neuroimage.usc.edu/brainstorm/Tutorials/TutExploreRecodings|tutorial #5 "Exploring the recordings"]], you can display a variety of 2D/3D maps of these recordings. Right-click on the node "Link to raw file" > MEG > ... {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=topo.gif|topo.gif|class="attachment"}} Remember that you can open a set of figures for a specific continuous file, save this configuration and re-open it with another file with the menu Window layout options > User setups. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=toolbarWindows.gif|topo.gif|class="attachment"}} === Cortical sources === As presented in tutorial #6 to #8, you can compute successively for this raw file: a head model, a noise covariance matrix, and an inverse model. Right-click on the folder "Subject01" to get all the menus. * '''Head model''': The goal of this tutorial is only to illustrate what you can do with the raw file viewer in Brainstorm, not to localize anything for real. Keep all the default options''' '''for the head model computation. * '''Noise covariance matrix''': Set as the baseline section the entire window before the first electric stimuation (the first occurrence of event "left"): [0, 4.6] seconds. Leave the other options to the default values. * '''Sources''': Keep the default options for the minimum norm method (wMNE). At the end you can see four new files in the database. Right-click on the source file associated with the "Link to raw file" and try the different visualization menus. Delete all these new files before you move on to the next tutorial, keep only the channel file and the "Link to raw file". {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=sourcesTree.gif|sourcesTree.gif|class="attachment"}} {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer?action=AttachFile&do=get&target=sourcesView.gif|sourcesView.gif|class="attachment"}} = C2. Detect and remove artifacts = ''Authors: Francois Tadel, Elizabeth Bock, John C Mosher, Sylvain Baillet'' It is common to have portions of recordings contaminated by events coming from the subject (eye blinks, movements, heartbeats, teeth clenching, implanted stimulators...) or from the environment (stimulation equipment, elevators, cars, trains, building vibrations...). Some occur at specific frequencies and can be removed using '''frequency filters''', as introduced in the first chapter of this tutorial. Some other artifacts have more complex frequency patterns but are well defined, reproducible and can be removed efficiently using '''Signal Space Projections''' ('''SSP'''). This tutorial shows how to apply this technique to correct for the cardiac and ocular artifacts. We are going to use the protocol '''TutorialRaw''' created in the previous tutorial [[http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer|Review continuous recordings and edit markers]]. If you have not followed this tutorial yet, please do it now. == Power line contamination == Notch filters are adapted to remove some well identified contaminations from oscillating systems, such as the power lines 50Hz or 60Hz. Here is an example of how to evaluate and correct fixed frequency artifacts. Important notes: 1. Usually you would prefer to apply the frequency filters '''before '''the SSP correction. 1. This approach is limited to CTF and Elekta-Neuromag recordings for now, other file formats will be developed on demand. An alternative approach for other file formats is described in the [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy|EEG/Epilepsy tutorial]]. === Identify the unwanted frequencies === Drag and drop the "Link to raw file" in the Process1 tab, click on the button [Run] to open the Pipeline editor window and select the process "'''Frequency > Power spectrum density (Welch)'''". This will estimate the power spectrum of the signal using the Welch method. Set the time window to process to '''[0,50]s''', and the window length to '''4 sec''', with an overlap of '''50%''', this will be more than enough to get a good estimate of the spectrum (average of the Fourier transform of 24 windows of 4800 samples each). Leave the other options to the default values. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=psdRun.gif|psdRun.gif|height="290",width="463",class="attachment"}} Double click on the "Power (MEG)" file that was created under the "Link to raw file" in the database explorer. This shows the estimate of the power spectrum for the first 50 seconds of the continuous file, for all the sensors, with a logarithmic scale. You can identify four peaks at the following frequencies: 60Hz, 120Hz, 180Hz and 300Hz. The first three are related with the power lines (acquisition in Canada, where the electricity is delivered at 60Hz, plus the harmonics), and are expected to be coming from pure sinusoidal components in the signal. The last one is an artifact of the low-pass filter at 300Hz that was applied on the recordings at the acquisition time. We are going to ignore this one, as it is probably more complex and spanning over several frequency bins. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=psdBefore.gif|psdBefore.gif|height="172",width="399",class="attachment"}} |
== Pre-processing == === Evaluate the recordings === * Drag and drop the "Link to raw file" into the Process1 list. * Select the process "'''Frequency > Power spectrum density'''", configure it as follows: <<BR>><<BR>> {{attachment:psdRun.gif||height="265",width="422"}} * Double-click on the PSD file to display it. It shows the estimate of the power spectrum for the first 50 seconds of the continuous file, for all the sensors, with a logarithmic scale. You can identify four peaks at the following frequencies: 60Hz, 120Hz, 180Hz and 300Hz. The first three are related with the power lines (acquisition in Canada, where the electricity is delivered at 60Hz, plus the harmonics). The last one is an artifact of the low-pass filter at 300Hz that was applied on the recordings at the acquisition time. <<BR>><<BR>> {{attachment:psdBefore.gif||height="187",width="396"}} |
| Line 291: | Line 48: |
| Leave the file "Link to raw file" in the Process1 list and now run "'''Pre-process > Notch filter'''". | * In Process1, keep the "Link to raw file" selected. * Run '''Pre-process > Notch filter''': Frequencies to remove = '''60, 120, ''''''180 Hz'''. <<BR>><<BR>> {{attachment:processSin.gif||height="252",width="307"}} * To evaluate the results of this process, select the new filtered file ('''"Raw | notch"''') and run again the process "'''Frequency > Power spectrum density'''". <<BR>><<BR>> {{attachment:processPsd2.gif||height="275",width="500"}} * You should observe a significant decrease of the contributions of the removed frequencies (60Hz, 120Hz, 180Hz) compared with the original spectrum. <<BR>><<BR>> {{attachment:psdAfter.gif||height="182",width="386"}} |
| Line 293: | Line 53: |
| . Enter the frequencies to remove ('''60Hz, 120Hz '''and '''180Hz''') and leave the other options to the default values. | === Heartbeats and blinks === Signal Space Projection (SSP) is a method for projecting the recordings away from stereotyped artifacts, such as eye blinks and heartbeats. |
| Line 295: | Line 56: |
| {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sinRemoval.gif|sinRemoval.gif|height="272",width="333",class="attachment"}} | * Double-click on the filtered continuous file to display all the '''MEG '''recordings. * Right-click on the link > '''ECG '''> Display time series, to look at the heartbeats. * Right-click on the link > '''EOG '''> Display time series, to look at the eye movements. * From the Artifacts menu in the Record tab, run the following detection processes: * '''Artifacts > Detect heartbeats:''' Select channel '''EEG057''', event name "cardiac". <<BR>><<BR>> {{attachment:detectEcg.gif||height="209",width="300"}} * Artifacts > '''Detect eye blinks:''' Select channel '''EEG058''', event name "blink". <<BR>><<BR>> {{attachment:detectEog.gif||height="207",width="300"}} * Artifacts > '''Remove simultaneous:''' To avoid capturing ocular artifacts in the cardiac SSP. <<BR>><<BR>> {{attachment:processRemoveSimult.gif||height="228",width="300"}} * Review the traces of ECG/EOG channels and make sure the events detected make sense. <<BR>><<BR>> {{attachment:events.gif}} * '''Artifacts >''' '''SSP: Heartbeats''': Event "cardiac", sensors="'''MEG'''". * Display the first three components: None of them can be clearly related to a cardiac component. This can have two interpretations: the cardiac artifact is not very strong for this subject and the influence of the heart activity over the MEG sensors is completely buried in the noise or the brain signals, or the characterization of the artifact that we did was not so good and we should refine it by selecting for instance smaller time windows around the cardiac peaks. Here, it's probably due to the subject's morphology. Some people generate strong artifacts in the MEG, others don't. * In this case, it is safer to '''unselect '''this "cardiac" category, rather than removing randomly spatial components that we are not sure to identify. <<BR>><<BR>> {{attachment:sspEcg.gif||height="189",width="651"}} * '''Artifacts >''' '''SSP: Eyeblinks''': Event "blink", sensors="'''MEG '''", use existing SSP. (select component #1) <<BR>><<BR>> {{attachment:sspEog.gif||height="185",width="501"}} |
| Line 297: | Line 69: |
| The operation creates a new "Link to raw file" entry in the database, pointing to a new CTF dataset in the same folder as the original continuous file. To check where this file was created: right-click on the file "'''Raw | notch(60Hz 120Hz 180Hz)'''" > '''File '''> '''Show in file explorer'''. This file has the exact same properties as the original file, including the SSP projectors, only the values of the MEG sensors were updated. | == Epoching and averaging == === Import the recordings === * Right-click on the filtered file "Raw | notch"''' > Import in dabase.''' <<BR>><<BR>> {{attachment:importMenu.gif}} * The following figure appears, and asks how to import these recordings in the Brainstorm database. <<BR>><<BR>> {{attachment:importOptions.gif||height="363",width="567"}} * '''Time window''': Time range of interest, keep all the time definition. * '''Split''': Useful to import continuous recordings without events. We do not need this here. * '''Events selection''': Check the "Use events" option, and select both '''Left''' and '''Right'''. * '''Epoch time''': Time instants that will be extracted before an after each event, to create the epochs that will be saved in the database. Set it to '''[-100, +300] ms''' * '''Use Signal Space Projections''': Use the active SSP projectors calculated during the previous pre-processing steps. Keep this option selected. * '''Remove DC Offset''': Check this option, and select: Time range: [-100, 0] ms. For each epoch, this will: compute the average of each channel over the baseline (pre-stimulus interval: -100ms to 0ms), then subtract it from the channel at all the times in [-100,+300]ms. * '''Resample recordings''': Keep this unchecked * '''Create a separate folder for each epoch type''': If selected, a new folder is created for each event type (here, it would create two folders "left" and "right"). This option is mostly for EEG studies with channel files shared across runs. In a MEG study, we usually recommend to use one channel file per run, and to import all the epochs from one run in the same folder. |
| Line 299: | Line 82: |
| {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sinRemovalDb.gif|sinRemovalDb.gif|height="144",width="326",class="attachment"}} | * Click on Import and wait. At the end, you are asked whether you want to ignore one epoch that is shorter than the others. This happens because the acquisition of the MEG signals was stopped less than 300ms after the last stimulus trigger was sent. Therefore, the last epoch cannot have the full [-100,300]ms time definition. This shorter epoch would prevent us from averaging all the trials easily. As we already have enough repetitions in this experiment, we can just ignore it. Answer '''Yes''' to this question to discard the last epoch. {{attachment:importShortEpoch.gif}} * At this stage, you should review all the trials (press F3 to jump to the next file), separately for the magnetometers and the gradiometers, to make sure that you don't have any bad trial that have been imported. If you find a bad trial: right-click on the file or on the figure > Reject trial. <<BR>><<BR>> {{attachment:importAfter.gif||height="178",width="534"}} |
| Line 301: | Line 85: |
| === Alternatives === If the notch filter is not giving satisfying result, you can use two other processes: |
|
| Line 304: | Line 86: |
| * '''Sinusoid removal''': This process can do a better job at removing precise frequencies by identifying the sinusoidal components and then subtracting them from the signals in time domain (not a frequency filter). * '''Band-stop filter''': This process can be useful for removing larger segments of the spectrum, in case the power line peaks are spread over numerous frequency bins or for suppressing other types of artifacts. |
|
| Line 307: | Line 87: |
| === Evaluate the results === Now run the same frequency analysis: process "'''Frequency > Power spectrum density (Welch)'''", time window = '''[0,50]s''', window length = '''4000ms''', overlap = '''50%'''. Double click on the new "Power (MEG)" file. As expected, the three first peaks due to the power lines are gone. |
=== Averaging === * Drag and drop all the left and right trials to the Process1 tab. * Run the process '''Average > Average files > By trial group (folder average)''': <<BR>><<BR>> {{attachment:processAverge.gif}} * Double-click on the Left and Right averages to display all the MEG sensors: <<BR>><<BR>>{{attachment:averageDisplay.gif}} |
| Line 310: | Line 92: |
| {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=psdAfter.gif|psdAfter.gif|height="177",width="411",class="attachment"}} | |
| Line 312: | Line 93: |
| You can run a few other pre-processing operations directly on the continuous files, including a band-pass filter. Frequency filters and sinusoid removals are operations that you should apply at very early stages of the analysis, before epoching the recordings. These operations do not perform well next to the beginning and the end of the signals, they may generate important artifacts. It is therefore much more accurate to filter the entire recordings from the original continuous file at once, rather than filtering small epochs after importing them in the database. From now on, we are only going to work on this clean file "Raw | notch(60Hz 120Hz 180Hz)". == Signal Space Projection == The Signal-Space Projection (SSP) is one approach to rejection of external disturbances. Here is a short description of the method by '''Matti Hämäläinen''', from the [[http://www.martinos.org/meg/manuals/MNE-manual-2.7.pdf|MNE 2.7 reference manual]], section 4.16. Unlike many other noise-cancellation approaches, SSP does not require additional reference sensors to record the disturbance fields. Instead, SSP relies on the fact that the magnetic field distributions generated by the sources in the brain have spatial distributions sufficiently different from these generated by external noise sources. Furthermore, it is implicitly assumed that the linear space spanned by the significant external noise patterns has a low dimension. Without loss of generality we can always decompose any ''n''-channel measurement '''b'''(''t'') into its signal and noise components as: . '''b'''(''t'') = '''b''',,''s'',,(''t'') ''+'' '''b''',,''n'',,(''t'') Further, if we know that '''b''',,''n'',,(''t'') is well characterized by a few field patterns '''b''',,''1'',,...'''b''',,''m'',,, we can express the disturbance as . '''b''',,''n'',,(''t'') = '''Uc''',,''n'',,(''t'') ''+'' '''e'''(''t'') , where the columns of '''U''' constitute an orthonormal basis for '''b''',,''1'',,...'''b''',,''m'',,, '''c''',,''n'',,(''t'') is an ''m''-component column vector, and the error term '''e'''(''t'') is small and does not exhibit any consistent spatial distributions over time,'' i.e.'', '''C''',,''e'',, = ''E''{'''ee'''^''T''^} = '''I'''. Subsequently, we will call the column space of '''U''' the noise subspace. The basic idea of SSP is that we can actually find a small basis set '''b''',,''1'',,...'''b''',,''m'',, such that the conditions described above are satisfied. We can now construct the orthogonal complement operator . '''P''',,⊥,, = '''I''' - '''UU'''^T^ and apply it to '''b'''(''t'') yielding . '''b'''(''t'') ≈ '''P''',,⊥,,'''b''',,''s'',,(''t'') , since '''P''',,⊥,,'''b''',,''n'',,(''t'') = '''P''',,⊥,,'''Uc''',,''n'',,(''t'') ≈ 0. The projection operator '''P''',,⊥,, is called the signal-space projection operator and generally provides considerable rejection of noise, suppressing external disturbances by a factor of 10 or more. The effectiveness of SSP depends on two factors: 1. The basis set '''b''',,''1'',,...'''b''',,''m'',, should be able to characterize the disturbance field patterns completely and 1. The angles between the noise subspace space spanned by '''b''',,''1'',,...'''b''',,''m'',, and the signal vectors '''b''',,''s'',,(''t'') should be as close to π/2 as possible. If the first requirement is not satisfied, some noise will leak through because '''P''',,⊥,,'''b''',,''n'',,(''t'') ≠ 0. If the any of the brain signal vectors '''b''',,''s'',,(''t'') is close to the noise subspace not only the noise but also the signal will be attenuated by the application of '''P''',,⊥,, and, consequently, there might by little gain in signal-to-noise ratio. Since the signal-space projection modifies the signal vectors originating in the brain, it is necessary to apply the projection to the forward solution in the course of inverse computations. For more information on the SSP method, please consult the following publications: * C. D. Tesche, M. A. Uusitalo, R. J. Ilmoniemi, M. Huotilainen, M. Kajola, and O. Salonen, "Signal-space projections of MEG data characterize both distributed and well-localized neuronal sources," Electroencephalogr Clin Neurophysiol, vol. 95, pp. 189-200, 1995. * M. A. Uusitalo and R. J. Ilmoniemi, "Signal-space projection method for separating MEG or EEG into components," Med Biol Eng Comput, vol. 35, pp. 135-40, 1997. == Identify the artifacts == The first step is to identify several repetitions of the artifact (the vectors '''b''',,''1'',,...'''b''',,''m'',,). We need to set markers in the recordings that indicate when the events that we want to correct for occur. To help with this task, it is recommended to always record with bipolar electrodes the activity of the eyes (electro-oculogram or EOG, vertical and horizontal), the heart (electro-cardiogram or ECG), and possibly other sources of muscular contaminations (electromyogram or EMG). In this example, we are going to use the ECG and vertical EOG traces to mark the cardiac activity and the eye blinks. Two methods can be used, manual or automatic. === EOG/ECG channels === * Select the protocol '''TutorialRaw''' created in the previous tutorial, and select the "Functional data" view (second button in the toolbar on top of the database explorer). * Double-click on the '''clean''' continuous recordings ("'''Raw | notch(60Hz 120Hz 180Hz)'''") to open the MEG recordings. * Set the length of the reviewed time window to '''3 seconds''' (in the Record tab, text box "Duration") * Right-click on the continuous recordings again > '''Misc > Display time series'''. . {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=tsMisc.gif|tsMisc.gif|height="148",width="577",class="attachment"}} * In this file the channel type "'''Misc'''" groups the two channels '''EEG057 '''('''ECG''', in green) and '''EEG058 '''('''vertical EOG''', in red). This configuration depends on the acquisition setup, and can be redefined afterwards in Brainstorm (right-click on the channel file > Edit channel file, and then change manually the string in the column Type for any channel). * Use the shortcuts introduced in the previous tutorial to adjust the vertical scale of this display: Shift+mouse wheel, +/- keys, or the buttons on the right side of the figure. * Then go further in time to see what is happening on these channels over the time: use the "'''>>>'''" buttons, or the associated shortcuts (read the tooltips of the button) * '''ECG''': On the green trace, you can recognize the very typical shape of the electric activity of the heart (P, QRS and T waves). This is a very good example, the signal is not always that clean. * '''EOG''': On the red trace, there is not much happening for most of the recordings except for a few bumps, typical of eye blinks, eg. at 33.590s. This subject is sitting very still and not blinking much. We can expect MEG recordings of a very good quality. * You can observe the contamination from a blink on the left-frontal sensors: move to '''33.590s''' (you can use the text boxes in the time panel, the Record tab, or the scrollbar in the figure), and select a subset of sensors (Shift+B, or right-click on the figure > Display setup > CTF LF) . {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=megEog.gif|megEog.gif|height="265",width="475",class="attachment"}} === Manual marking === Create a new category of markers "'''blink_manual'''", using the menu Events > Add group. Select this new group. Review the file, and mark the peaks you observe on the vertical EOG trace, using the '''Ctrl+E''' keyboard shortcut. Do that for a few eye blinks. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=markEog.gif|markEog.gif|height="174",width="496",class="attachment"}} You could repeat the same operation for all the blinks, then for all the ECG peaks and jump to the next chapter of the tutorial and compute the SSP. It would be uselessly time consuming, as there is a process that does it for you automatically. However, it is good to remember how to do it manually because you may face some cases where you don't have clean ECG/EOG, or if you want to correct for another type of artifact. === Automatic detection: EOG === In the Record tab, select the menu: '''SSP > Detect eye blinks'''. It automatically opens the pipeline editor, with the process "Detect eye blinks" selected: * '''Channel name''': Name of the channel that is used to perform the detection. Select or type "'''EEG058'''" as it is the name of the EOG channel * '''Time window''': Time range that the algorithm should scan for the selected artifact. Leave the default values to process the entire file. * '''Event name''': Name of the event group that is created for saving all the detected events. Leave the default "blink". {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspMenu.gif|sspMenu.gif|class="attachment"}} {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=detectEog.gif|detectEog.gif|height="232",width="334",class="attachment"}} Click on Run. After the process stops, you can see two new event categories "'''blink'''" and "'''blink2'''" in the Record tab. You can review a few of them, to make sure that they really indicate the EOG events. In the Record tab, click on the "blink" event category, then on a time occurrence to jump to it in the MEG and Misc time series figures. Two types of events are created because this algorithm not only detects specific events in a signal, it also classifies them by shape. If you go through all the events that were detected in the two categories, you would see that the "blink" are all round bumps, typical of the '''eye blinks'''. In the category "blink2", the morphologies don't look as uniform; it mixes small blinks, and ramps or step functions followed by sharp drops that could indicate '''eye saccades'''. The saccades can be observed on the vertical EOG, but if you want a better characterization of them you should also record the horizontal EOG. The detection of the saccades should be performed with a different set of parameters, using the process "Detect custom events", introduced later in this chapter. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=detectEogDone.gif|detectEogDone.gif|class="attachment"}} === Automatic detection: ECG === Now do the same thing for the heartbeats. In the Record tab, select the menu "'''SSP > Detect heartbeats'''". Configure the process to use the channel '''EEG057''' (name of the ECG channel), and leave the other options to the default values. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspMenu1.gif|sspMenu1.gif|class="attachment"}} {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=detectEcg.gif|detectEcg.gif|height="228",width="330",class="attachment"}} Click on Run. After the process stops, you can see a new event category "'''cardiac'''" in the Record tab, with 346 occurrences. You can check a few of them, to make sure that the "cardiac" markers really indicate the ECG peaks, and that there are not too many peaks that are skipped. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=detectEcgDone.gif|detectEcgDone.gif|class="attachment"}} === Automatic detection: Custom === These two previous processes are shortcuts for a generic process "'''Detect custom events'''". We are not going to use it here, but it is interesting to introduce it to understand how the blinks and heartbeats detection work. The logic is the following: * The channel to analyze is read from the continuous file, for a given time window. * '''Frequency band''': The signal is filtered in a frequency band where the artifact is easy to detect. For EOG: 1.5-15Hz ; for ECG: 10-40Hz. * '''Threshold''': An event of interest is detected if the absolute value of the filtered signal value goes over a given number of times the standard deviation. For EOG: 2xStd, for ECG: 4xStd * '''Minimum duration between two events''': If the filtered signal crosses the threshold several times in relation with the same artifact (like it would be the case for muscular activity recordings on an EMG channel), we don't want to trigger several events but just one at the beginning of the activity. This parameter would indicate the algorithm to take only the maximum value over the given time window; it also prevents from detecting other events immediately after a successful detection. For the ECG, this value is set to 500ms, because it is very unlikely that the heart rate of the subject goes over 120 beats per minute. * '''Ignore the noisy segments''': If this option is selected, the detection is not performed on the segments that are much noisier than the rest of the recordings. * '''Enable classification''': If this option is selected, the events are classified by shape, based on correlation measure. In the end, only the categories that have more than 5 occurrences are kept, all the other successful detections are ignored. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=detectCustom.gif|detectCustom.gif|height="459",width="329",class="attachment"}} == Calculate the projectors == === Methodology === We have now 346 examples of heartbeats, 18 examples of blinks, and 12 examples of saccades. These are sufficient numbers of repetitions to calculate reliable signal-space projections. The logic of the computation is the following: 1. Take a small time window around each marker to capture the full effect of the artifact, plus some clean brain signals before and after. The default time window is [-200,+200]ms for eye blinks, and [-40,+40]ms for the heartbeats. 1. Filter the MEG or EEG signals in a frequency band of interest, in which the artifact is the most visible. 1. Concatenate all these time blocks into a big matrix '''A''' = ['''b''',,''1'',,, ..., '''b''',,''m'',,] 1. Compute the singular value decomposition of this matrix '''A''': ['''U''','''S''','''V'''] = '''svd'''('''A''', 'econ') 1. The singular vectors '''U'''i with the highest singular values '''S'''i are an orthonormal basis of the artifact subspace that we want to subtract from the recordings. The software selects by default the first vector only but it is possible to redefine interactively the selected components. 1. Calculate the projection operator:''' P''',,⊥i,, = '''I''' - '''U'''i'''U'''i^T^ 1. Apply this projection on the MEG or EEG recordings '''F''': '''F''' = '''P''',,⊥i,,'''F''' 1. The process has to be repeated '''separately '''several times: * for each sensor type (EEG, MEG magnetometers, MEG gradiometers) and * for each artifact, starting with the one that leads to the strongest contamination in amplitude on the sensors (in the case: the eye blinks) * If you have difficulties removing one artifact or the other, you may try to process them in a different order. You may also try removing some of the artifact markers in the case of co-occurring artifacts. If a lot of the blinks happen at the same time as heartbeats, you may end up calculating projectors that mix both effects, but that do not remove efficiently one or the other. In this case, remove all the markers that happen in the segments contaminated by multiple artifacts. Steps #1 to #5 are automatically done by the processes "SSP > Compute SSP" in the Record tab: the results, the vectors '''U'''i, are saved in the database link ("Link to raw file") and in the channel file. Steps #6 and #7 are calculated on the fly when reading a block of recordings from the continuous file: when using the raw viewer, running a process a process on the continuous file, or importing epochs in the database. Step #8 is the manual control of the process. Take some time to understand what you are trying to remove and how to do it. Never trust blindly any fully automated artifact cleaning algorithm, always check manually what is removed from the recordings, and do not give up if the first results are not satisfying. === Recommended procedure === We have multiple types of artifact to correct for and it can be tricky to know what artifact to correct first. If you correct for artifact A and then for artifact B, you may get different results than if you correct first B and then A. We propose here a default order for the two artifacts that we have in all the recordings: the blinks and the heartbeats. If two artifacts happen at the same time, the SSP projectors calculated for the blink may contain some of the heartbeat and vice versa. When trying to remove the second artifact, we might not be able to isolate it clearly anymore. To avoid this behavior, we should try to avoid having overlapping artifacts. Because the subject's heart is beating at least once per second, there is a high chance to record a heartbeat next to any eye blink. Therefore a significant number of the blinks will be contaminated with heartbeats. If we decide to remove all the blink events that include some heart activity, we may not have enough blink samples left to build correct SSP projectors. To isolate correctly these two common contaminations we recommend the following procedure: 1. Remove all the cardiac peaks that are occurring next to a blink (we can remove half of the heartbeats, we will still have enough data for calculating the projectors). 1. Calculate the SSP to remove the '''cardiac '''artifact. 1. Calculate the SSP to remove the '''eye blinks''', based on the recordings already corrected for the cardiac artifact. === Remove simultaneous events === If you closed the file viewer, open the MEG recordings again: . double click on the clean file "Raw | notch(60Hz 120Hz 180Hz)". In the Record tab, select the menu "'''SSP > Remove simultaneous'''". Set the options: * Remove events named: "'''cardiac'''" * When too close to events: "'''blink'''" * Minimum delay between events: '''250ms''' {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspMenuRemove.gif|sspMenuRemove.gif|class="attachment"}} {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=removeSimult.gif|removeSimult.gif|height="234",width="309",class="attachment"}} After executing this process, the number of "cardiac" events goes from '''346''' to '''333'''. The deleted heartbeats were all less than 250ms away from a blink. === Compute SSP: Heartbeats === In the Record tab, select the menu "'''SSP > Compute SSP: Heartbeats'''". Set the options: * '''Event name''': Name of the event to use to calculate the projectors, enter "'''cardiac'''" * '''Sensor types''': Type of sensors for which the projection should be calculated ("'''MEG'''"). It doesn't matter if there are other sensor types indicated in the text box: the sensor names or types that do not exist in the recordings are just ignored. Note that you will always get better results if you process the different types of sensors separately. If you have MEG and EEG in the same file, you should run twice this process, once for the EEG only and once for the MEG only. Same thing when processing Elekta-Neuromag recordings, you should process separately the magnetometers (MEG MAG) and the gradiometers (MEG GRAD). * '''Compute using existing SSP projectors''': You have the option to calculate the projectors from the raw recordings, or from the recordings filtered with the previously computed SSP projectors. Unless you have a good reason for not considering the existing SSP projectors, you should always select this option. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspEogFile.gif|sspEogFile.gif|class="attachment"}} {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspEcg.gif|sspEcg.gif|height="212",width="309",class="attachment"}} After the computation is done, a new figure is displayed, that lets you select the active projectors. On the left you have the projector categories (matrix '''U''') where each row represents the result of an execution of a "Compute SSP " process (usually one for each sensor type and each artifact). On the right, you can activate independently the different components for the selected projector category. Each entry represents the projector created from a column vector '''U'''i (singular vector of the SVD decomposition), and the percentage is the singular value for this vector, normalized for this decomposition (percentage = '''S'''i / sum('''S'''i)). When a projector is selected, it means that it is applied to the recordings on the fly when reading from the continuous file, ie. that the corresponding spatial component is removed from the recordings. The percentage indicates the amount of signal that was captured by this projector during the decomposition. The highest it is, the more the projector is representative of the artifact recordings that were used to calculate it. In the good cases, you would typically see one to three components with values that are significantly higher that the others. By default, the software selects '''the first component''' and leaves all the others unselected. Do not trust this automatic selection, you should always review manually the components that you want to remove. A list of buttons is available to manipulate the projector categories: load, save, rename, delete, and '''display component topography'''. The last one is one of the most useful tool to understand these results. Select the category ("cardiac"), click on the first component, then click on the toolbar button to display its topography. Do it again for components #2 and #3 (the components don't need to be active for that). The other button [No interp] displays the same results without the magnetic field smoothing that is applied by default to MEG topographies. It may help understand some difficult cases. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspEcgCheck.gif|sspEcgCheck.gif|height="207",width="390",class="attachment"}} {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspEcgComp.gif|sspEcgComp.gif|class="attachment"}} None of these topographies can be clearly related to a cardiac component. This can have two interpretations: the cardiac artifact is not very strong for this subject and the influence of the heart activity over the MEG sensors is completely buried in the noise or the brain signals, or the characterization of the artifact that we did was not so good and we should refine it by selecting for instance smaller time windows around the cardiac peaks. Here, it's probably due to the subject's morphology. Some people generate strong artifacts in the MEG, some don't. In this case, it is safer to '''unselect '''this "cardiac" category, rather than removing randomly spatial components that we are not sure to identify. === Compute SSP: Eye blinks === Let's try the same thing with the eye blinks. Select the process "'''SSP > Compute SSP: Eye blinks'''". Run it on the event type "blink", that indicate the peaks of the EOG signal. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspEogFile.gif|sspEogFile.gif|class="attachment"}} {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspEog.gif|sspEog.gif|height="214",width="307",class="attachment"}} In this specific case, there is one value that is much higher than the others (21%). The first component is most likely a good representation of the artifact we are trying to remove. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspEogSelect.gif|sspEogSelect.gif|height="214",width="384",class="attachment"}} Select the cardiac category, and display the topographies for the first three components. Component #1 is clearly related with the ocular activity occurring during the blinks, the others are not as clear. Conclusion: the default proposition was good (only component #1 selected). Setting this first projector as active removes from the MEG signals the contributions related with the spatial topography we can see in the first figure, ie. a spatial distribution of sensor values related specifically to the eye blinks. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspEogComp.gif|sspEogComp.gif|class="attachment"}} While the recordings are displayed in the file viewer, the selection of the active projectors interactively updates the recordings. Go to the first detected "blink" event ('''t=33.600s'''), and select the left frontal sensors ('''Shift+B''' or right-click > Display setup > CTF LF). Then play with the checkboxes to change the active projectors, and see how well this first component of the blink category removes the ocular artifact. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspEogMeg.gif|sspEogMeg.gif|class="attachment"}} Select the projector category "blink", select the first component, and click on Save, to validate your changes. Starting from now, all the successive operations you apply on this file will integrate this eye blink correction, including the calculation of other projectors. After you closed this window, you can get back to it by selecting the menu "'''SSP > Select active projector'''" in the Record tab, and change again the selection of active projectors. We are not going to calculate the SSP for the other category of events identified on the EOG, the group "'''blink2'''". The events classified in this group could represent eye saccades or other types of blinks, it is not very clear that they correspond to the same physiological event. Most of them do not seem to create any obvious contamination on the MEG recordings, there is probably no need to consider them now. === Compute SSP : Generic === The calculation of the SSP for the heartbeats and the eye blinks are shortcuts to a more generic algorithm "'''Compute SSP: Generic'''". We do not need it here, but the illustration of its properties is interesting to understand the correction of ocular and cardiac artifacts: * '''Event name''': Markers that are used to characterize the artifact * '''Time window''': Time segment to consider before and after each event marker. We want this time window to be longer than the artifact effect itself, in order to have a large number of time samples representative of a normal brain activity. This helps the SVD decomposition to separate the artifact from the ongoing brain activity. * '''Frequency band''': Definition of the band-pass filter that is applied to the recordings before calculating the projector. Usually you would use the same frequency band as we used for the detection, but you may want to try to refine this parameter if the results are not satisfying. * '''Sensor types or names''': List of sensors for which to calculate the projector. You can get better results if you process one sensor type at a time. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=sspGeneric.gif|sspGeneric.gif|height="328",width="330",class="attachment"}} === Troubleshooting === You have calculated the SSP as indicated here but you don't get any good results. No matter what you do, the topographies don't look like the targeted artifact. You can try the following: * Review one by one the events indicating the artifacts, remove the ones that are less clear or that occur close to another artifact. * Select or unselect the option "Compute using existing SSP". * Use the process "Compute SSP: Generic" and modify some parameters: * Reduce the time window around the peak of the artifact. * Use a narrower frequency band: especially the EOG, if the projectors capture some of the alpha oscillations, you can limit the frequency band to [1.5 - 9] Hz. * Change the method: Average / SSP. * If you have multiple acquisition runs, you may try to use all the artifacts from all the runs rather than processing the files one by one. For that, use the Process2 tab instead of Process1. Put the "Link to raw file" of all the runs on both side, Files A (what is used to compute the SSP) and Files B (where the SSP are applied). == Evaluation == === Eye blinks === One efficient way of representing the impact of this artifact correction is to epoch the recordings around the artifacts before and after the correction. Use the same time window as the one used in the SSP options around each marker, and average all the segments of recordings. These operations are detailed in the next tutorial, we are just presenting the results. You don't need to reproduce them at this time, just remember that it is doable, in case you need it with your own data. This is what the database would look like after these operations: {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=blinkDb.gif|blinkDb.gif|height="263",width="283",class="attachment"}} The following image represents the file '''"blink_uncorrected / Avg: blink"''', which is the average of the 18 identified blinks '''before''' the SSP correction, [-200,+200]ms around the "blink" events. The time series, the 2D topography at the peak (t = 0ms), and the mapping on the cortex of these fields, using the basic inverse model calculated in the previous tutorial. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=blinkBefore.gif|blinkBefore.gif|height="177",width="533",class="attachment"}} The next image represents the file '''"blink_ssp / Avg: blink"''', which is the exact same thing, but '''after '''the SSP correction. The artifact is gone. If you do this and you can still see some clear evoked component in the time series figure, the SSP correction was not efficient: the artifact is not properly identified, or you should select different components using the "Select active projectors" window. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=blinkAfter.gif|blinkAfter.gif|height="177",width="533",class="attachment"}} === Heartbeats === We can do the same thing with the heartbeats: epoch [-40,+40]ms around the "cardiac" events, average the trials, and review the event-related fields at three instants: The peak of the '''P wave''' (t = '''-30ms'''): {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=cardiacP.gif|cardiacP.gif|height="178",width="534",class="attachment"}} The peak of the '''QRS complex''' (t = '''0ms'''): {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=cardiacQRS.gif|cardiacQRS.gif|height="178",width="534",class="attachment"}} The peak of the '''T wave''' (t = '''25ms'''): {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp?action=AttachFile&do=get&target=cardiacT.gif|cardiacT.gif|height="178",width="534",class="attachment"}} These peaks may look big, but they are in fact much smaller (300 fT for the average of 346 repetitions) than what we observed for the eye blinks (1500 fT for 18 repetitions). You can notice that none of these topographies are similar to the components we obtained after calculating the SSP for the cardiac artifact. We don't know how to correct this artifact, it doesn't look too bad in terms of recordings contamination, so we just leave it uncorrected. = C3. Epoching and averaging = ''Authors: Francois Tadel, Elizabeth Bock, John C Mosher, Sylvain Baillet'' This tutorial fills the gap between the previous tutorial (review and clean continuous recordings), and the introduction tutorials (source analysis of evoked responses). It explains how to epoch the recordings, do some more pre-processing on the single trials, and then calculate their average. For this tutorial, we are going to use the protocol '''TutorialRaw''' created in the two previous tutorials: [[http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawViewer|Review continuous recordings and edit markers]] and [[http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawSsp|Artifact cleaning]]. If you have not followed these tutorials yet, please do it now. == Import in database == The raw file viewer provides a rapid access to the recordings, but most of operations cannot be performed directly on the continuous files: most of the pre-processing functions, averaging, time-frequency analysis and statistical tests can only be applied on blocks of data that are saved in the database (ie. "imported"). After reviewing the recordings and editing the event markers, you have to "import" the recordings in the database to go with further analysis. Right-click on the file with power line correction:''' Raw | notch(60Hz 120Hz 180Hz) > Import in dabase''' {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=importMenu.gif|importMenu.gif|class="attachment"}} Warning: If you right-click on the subject node > Import EEG/MEG, and select the tutorial .ds dataset, you would be able to import blocks of the continuous file in the database, but you would not have access to the modified events list or the SSP operators. Therefore, you would not import data cleaned of ocular and cardiac artifacts. The modified events list and the signal space projectors are saved only in the "Link to raw file" in the Brainstorm database, not in the initial continuous file. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=importOptions.gif|importOptions.gif|class="attachment"}} Set the import options as they are represented in this figure: * '''Time window''': Time range of interest. We are interested by all the stimulations, so do not change this parameter; the default values always represent the entire file. * '''Split''': Useful to import continuous recordings without events, to import successive chunks of the same duration. We do not need this here. * '''Events selection''': Check the "Use events" option, and select both "left" and "right". The number in the parenthesis represents the number of occurrences of this event in the selected time window (would change if you modify the time definition on top of the figure) * '''Epoch time''': Time segment that is extracted around each marker and saved in the database. Set it to [-100, +300] ms * '''Use Signal Space Projections''': Use the active SSP projectors calculated during the previous pre-processing steps. Keep this option selected. * '''Remove DC Offset''': Check this option, and select: Time range: [-100, 0] ms. For each epoch, this will: compute the average of each channel over the baseline (pre-stimulus interval: -100ms to 0ms), and subtract it from the channel at every time instants (full epoch interval: [-100,+300]ms). This option removes the baseline value of each sensor, ie. the continuous (DC) offset that is added permanently on top of the recordings of interest. In MEG, the sensors record variations around a somewhat arbitrary level, therefore this operation is always needed, unless it was already applied during one of the pre-processing steps. Note that a high-pass filter with a very low frequency (for instance 0.3Hz) can replace efficiently this DC correction. If a high-pass filter has already been applied to the recordings, you may want to unselect this option. * '''Resample recordings''': Keep this unchecked * '''Create a separate folder for each epoch type''': If selected, a new folder is created for each event type (here, it will create two folders in the database: "left" and "right"). If not selected, all the epochs are saved in a new folder, the same one for all the events, that has the same name as the initial raw file. Click on Import and wait. At the end, you are asked whether you want to ignore one epoch that is shorter than the others. This happens because the acquisition of the MEG signals was stopped less than 300ms after the last stimulus trigger was sent. Therefore, the last epoch cannot have the full [-100,300]ms time definition. This shorter epoch would prevent us from averaging all the trials easily. As we already have enough repetitions in this experiment, we can just ignore it. Answer '''Yes''' to this question to discard the last epoch. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=importShortEpoch.gif|importShortEpoch.gif|class="attachment"}} Two new conditions containing two groups of trials appeared in the database. To expand a group of trials and get access to the individual epochs: double-click on it or click on the "+" next to it. The SSP projectors calculated in the previous tutorial were applied on the fly when reading from the continuous file. These epochs are clean from eye blinks and power line contamination. All the files available in the ''(Common files)'' folder are also shared for these new folders "left" and "right". == Review the individual trials == Double-click on the first trial for the "left" condition. Then right-click on the figure > Navigator > '''Next data file''', or use the keyboard shortcut '''F3 '''to jump to the next trial. This way you can quickly review all the trials, to make sure that there is no obvious problem in the recordings. If you haven't reviewed manually all the recordings in the continuous mode, and marked all the bad segments, it is a good time to do it now. To mark a trial as bad manually, you have three methods: * Right-click on the trial file in the database > '''Reject trial''' * Right-click on the figure > '''Reject trial''' * Use the keyboard shortcut '''Control+B''' * To set all the trials back as good in a group: right-click on the trials group or the condition > Accept bad trials. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=rejectManual.gif|rejectManual.gif|class="attachment"}} When a trial is tagged as bad, its icon in the database explorer shows a red mark. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=rejectTree.gif|rejectTree.gif|class="attachment"}} All the bad trials are going to be ignored in the rest of the analysis, because they are ignored by the Process1 and Process2 tabs. If you drag and drop the 101 left trials in the Process1 list, with one trial marked as bad, the summary of the selected files on top of the tab would show only 100 data files. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=rejectProcess.gif|rejectProcess.gif|class="attachment"}} To learn how to mark only individual channels as bad instead of the entire trial, please go back to the introduction tutorial [[http://neuroimage.usc.edu/brainstorm/Tutorials/TutExploreRecodings|Exploring the recordings]]. The process "'''Artifacts > Detect bad channels: peak-to-peak'''" can help you detect some bad trials based on a peak-to-peak amplitude threshold. Drag and drop all the trials from the '''left''' condition in the Process1 list, and select this process. For each trial, it detects the channels that have a peak-to-peak amplitude (maximum value - minimum value) that is above a rejection threshold (too noisy) or below a rejection threshold (no signal). You can define different thresholds for different channel types. The options are: * '''Time window''': Time range on which the peak-to-peak amplitude is calculated on each trial * '''Thresholds''': Rejection and detection thresholds for each sensor type. For CTF recordings, use the "MEG gradio" category, and ignore the "/cm (x 0.04)" indication, that is valid only for Neuromag systems. Set the '''MEG gradio''' thresholds to '''[0, 2000] fT''', to detect the MEG channels that have a peak-to-peak amplitude superior to 2000fT. * '''Reject only the bad channels''': This would tag as bad only the detected channels, but would not tag the trial itself as bad. * '''Reject the entire trial''': This would tag as bad the trial if there is at least one channel file identified as bad. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=rejectOptions.gif|rejectOptions.gif|class="attachment"}} For this current dataset, the quality of the recordings is remarkably good, we don't need to mark any trial as bad. So right-click on the group of "left" trials > Accept trials, to set them all as good. == Averaging == Drag and drop the two lists of trials (or the two condition folders) in the Process1 list, and run the process "'''Average > Average files'''", with the option "'''Average by condition (subject average)'''". This will group the trials by conditions, and calculate one average per condition and subject. The option "By condition (grand average)" would average each condition for all the subjects together, but as there is only one subject in this case, the result would be same. The other options would generate one average file per subject ("by subject"), one average file per group of trials and per subject ("by trial group"), or only one average no matter what the input is ("everything"). The function to apply is the regular arithmetic average. The option "'''Keep all the events from the individual epochs'''" would group all the event markers present in all the epochs and save them in the new averaged file. It can be useful to check the relative position of the artifacts or the subject responses, or quickly detect some unwanted configuration such as a subject who would constantly blink right after a visual stimulus. We don't really need this here, leave this option '''unselected'''. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=avgOptions.gif|avgOptions.gif|class="attachment"}} It creates two files: "'''Avg: left'''" and "'''Avg: right'''". {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=avgDb.gif|avgDb.gif|class="attachment"}} Double-click on the "'''Avg: Left'''" file to display the MEG recordings for this file (or right-click > MEG > display time series). It shows a very typical an clean evoked response, with a very high signal-to-noise ratio. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=avgTs.gif|avgTs.gif|class="attachment"}} Just for the records, this is how it would look like if we had selected the option "Keep all the events from the individual epochs". The summary of the markers of all the epochs is: 37 heartbeats and 2 blinks. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=avgTsEvents.gif|avgTsEvents.gif|class="attachment"}} |
|
| Line 662: | Line 110: |
== Filters for visualization == As introduced in the previous tutorials, you can add a visualization filter to make the traces and the topographies look smoother. We suggest in this case a low-pass filter at 120Hz, because it shows very smooth traces, but all the waves we are interested in are still clearly visible in the MEG time series figure. {{http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawAvg?action=AttachFile&do=get&target=onlineFilter.gif|onlineFilter.gif|class="attachment"}} |
|
| Line 727: | Line 170: |
| All the operations that have been introduced in the last three tutorials can be reproduced easily from a Matlab script. This is illustrated in the tutorial [[http://neuroimage.usc.edu/brainstorm/Tutorials/TutRawScript|Full analysis with one script]]. | The following script from the Brainstorm distribution reproduces the analysis presented in this tutorial page: '''brainstorm3/toolbox/script/''''''tutorial_raw.m ''' |
| Line 729: | Line 172: |
| This script is available in the Brainstorm distribution: '''brainstorm3/toolbox/script/tutorial_raw.m ''' | <<HTML(<div style="border:1px solid black; background-color:#EEEEFF; width:720px; height:500px; overflow:scroll; padding:10px; font-family: Consolas,Menlo,Monaco,Lucida Console,Liberation Mono,DejaVu Sans Mono,Bitstream Vera Sans Mono,Courier New,monospace,sans-serif; font-size: 13px; white-space: pre;">)>><<EmbedContent("http://neuroimage.usc.edu/bst/viewcode.php?f=tutorial_raw.m")>><<HTML(</div >)>> |
MEG median nerve tutorial (CTF)
Authors: Francois Tadel, Elizabeth Bock, John C Mosher, Sylvain Baillet
The dataset presented in this tutorial was used in the previous generation of introduction tutorials. We kept it on the website as an additional illustration of data analysis, and as a comparison with median nerve experiments recorded with other MEG systems (Elekta-Neuromag, Yokogawa). No new concepts are presented in this tutorial. For in-depth explanations of the interface and theoretical foundations, please refer to the introduction tutorials.
Contents
License
This tutorial dataset (MEG and MRI data) remains a property of the MEG Lab, McConnell Brain Imaging Center, Montreal Neurological Institute, McGill University, Canada. Its use and transfer outside the Brainstorm tutorial, e.g. for research purposes, is prohibited without written consent from the MEG Lab.
If you reference this dataset in your publications, please acknowledge its authors (Elizabeth Bock, Esther Florin, Francois Tadel and Sylvain Baillet) and cite Brainstorm as indicated on the website. For questions, please contact us through the forum.
Presentation of the experiment
Experiment
- One subject, one acquisition run of 6 minutes
- Subject stimulated using Digitimer Constant Current Stimulator (model DS7A)
- The run contains 200 electric stimulations randomly distributed between left and right:
- 102 stimulations of the left hand
- 98 stimulations of the right hand
- Inter-stimulus interval: jittered between [1500, 2000]ms
Stimuli generated using PsychToolBox on Windows PC (TTL pulse generated with the parallel port connected to the Digitimer via the rear panel BNC)
MEG acquisition
Acquisition at 1200Hz, with a CTF 275 system, subject in seating position
- Recorded at the Montreal Neurological Institute in November 2011
- Anti-aliasing low-pass filter at 300Hz, files saved with the 3rd order gradient
- Recorded channels (302):
- 1 Stim channel indicating the presentation times of the stimuli: UPPT001 (#1)
- 26 MEG reference sensors (#3-#28)
- 272 MEG axial gradiometers (#29-#300)
- 1 ECG bipolar (#301)
- 1 vertical EOG bipolar (#302)
- 1 Unused channels (#2)
1 dataset: subj001_somatosensory_20111109_01_AUX-f.ds
Stimulation delays
Delay #1: 4 samples @ 1200Hz = 3.3ms. The CTF MEG system has an anti-aliasing filter (1/4 the sampling rate) applied at the time of recording. This filter is applied only to the MEG channels and not to the stimulus channels. Therefore the MEG has a 4 sample delay when compared with the stimulus channel.
Head shape and fiducial points
3D digitization using a Polhemus Fastrak device (subj00111092011.pos)
Position of the anatomical references used for these recordings:
The position of the CTF head localization coils, pictures available in file fiducials.jpg- Around 110 head points distributed on the hard parts of the head (no soft tissues)
Subject anatomy
- Subject with 1.5T MRI
Processed with FreeSurfer 5.2
Download and installation
Requirements: You have already followed all the basic tutorials and you have a working copy of Brainstorm installed on your computer.
Go to the Download page of this website, and download the file: sample_raw.zip
- Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder).
- Start Brainstorm (Matlab scripts or stand-alone version).
Select the menu File > Create new protocol. Name it "TutorialRaw" and select the options:
No, use individual anatomy,
No, use one channel file per run.
Import the anatomy
- Create a new subject Subject01.
Right-click on the subject node > Import anatomy folder:
Set the file format: "FreeSurfer folder"
Select the folder: sample_raw/anatomy
- Number of vertices of the cortex surface: 15000 (default value)
Click on the link "Click here to compute MNI transformation".
- Set the 3 required fiducial points (indicated in MRI coordinates):
- NAS: x=127, y=212, z=123
- LPA: x=55, y=124, z=119
- RPA: x=200, y=129, z=114
At the end of the process, make sure that the file "cortex_15000V" is selected (downsampled pial surface, that will be used for the source estimation). If it is not, double-click on it to select it as the default cortex surface.

Link the recordings
- Switch to the "functional data" view (middle button in the toolbar above the database explorer).
Right click on the Subject01 > Review raw file:
Select the file format: MEG/EEG: CTF
Select the folder: sample_raw/Data/subj001_somatosensory_20111109_01_AUX-f.ds
Refine the registration with the head points: YES.
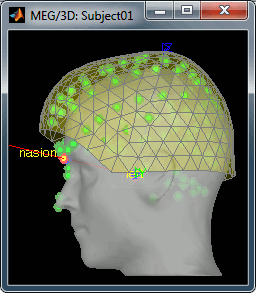
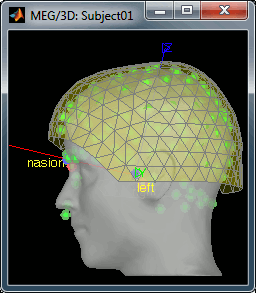
Pre-processing
Evaluate the recordings
- Drag and drop the "Link to raw file" into the Process1 list.
Select the process "Frequency > Power spectrum density", configure it as follows:
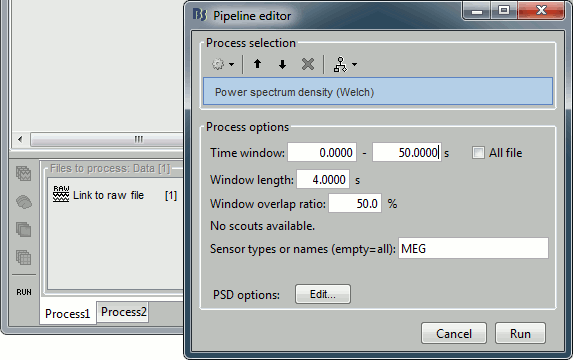
Double-click on the PSD file to display it. It shows the estimate of the power spectrum for the first 50 seconds of the continuous file, for all the sensors, with a logarithmic scale. You can identify four peaks at the following frequencies: 60Hz, 120Hz, 180Hz and 300Hz. The first three are related with the power lines (acquisition in Canada, where the electricity is delivered at 60Hz, plus the harmonics). The last one is an artifact of the low-pass filter at 300Hz that was applied on the recordings at the acquisition time.
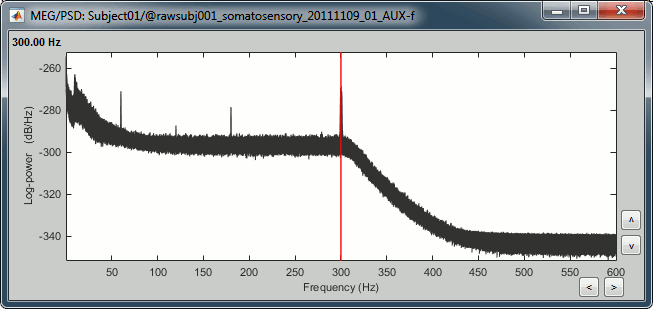
Remove: 60Hz and harmonics
- In Process1, keep the "Link to raw file" selected.
Run Pre-process > Notch filter: Frequencies to remove = 60, 120, 180 Hz.
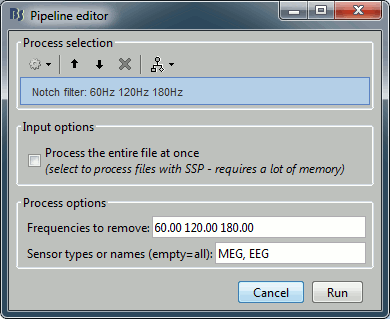
To evaluate the results of this process, select the new filtered file ("Raw | notch") and run again the process "Frequency > Power spectrum density".
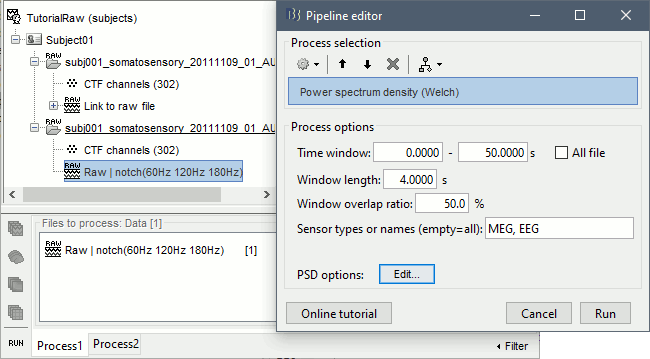
You should observe a significant decrease of the contributions of the removed frequencies (60Hz, 120Hz, 180Hz) compared with the original spectrum.
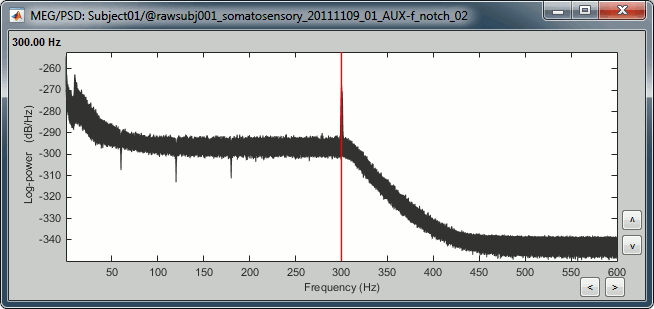
Heartbeats and blinks
Signal Space Projection (SSP) is a method for projecting the recordings away from stereotyped artifacts, such as eye blinks and heartbeats.
Double-click on the filtered continuous file to display all the MEG recordings.
Right-click on the link > ECG > Display time series, to look at the heartbeats.
Right-click on the link > EOG > Display time series, to look at the eye movements.
- From the Artifacts menu in the Record tab, run the following detection processes:
Artifacts > Detect heartbeats: Select channel EEG057, event name "cardiac".
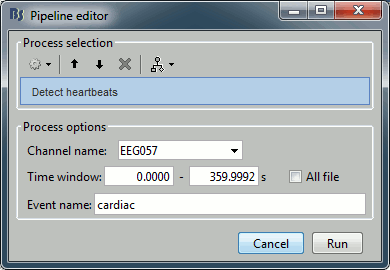
Artifacts > Detect eye blinks: Select channel EEG058, event name "blink".
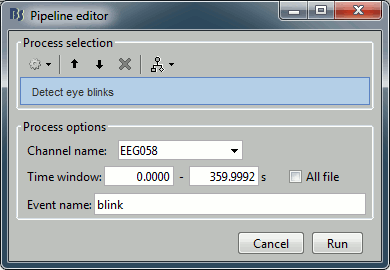
Artifacts > Remove simultaneous: To avoid capturing ocular artifacts in the cardiac SSP.
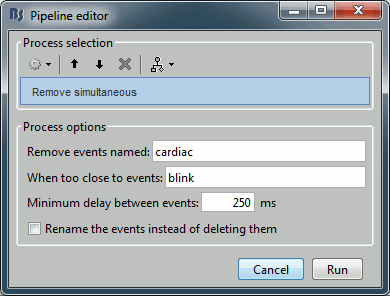
Review the traces of ECG/EOG channels and make sure the events detected make sense.
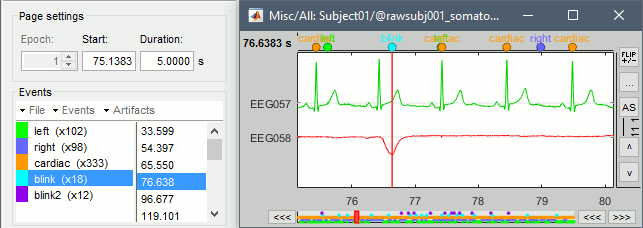
Artifacts > SSP: Heartbeats: Event "cardiac", sensors="MEG".
- Display the first three components: None of them can be clearly related to a cardiac component. This can have two interpretations: the cardiac artifact is not very strong for this subject and the influence of the heart activity over the MEG sensors is completely buried in the noise or the brain signals, or the characterization of the artifact that we did was not so good and we should refine it by selecting for instance smaller time windows around the cardiac peaks. Here, it's probably due to the subject's morphology. Some people generate strong artifacts in the MEG, others don't.
In this case, it is safer to unselect this "cardiac" category, rather than removing randomly spatial components that we are not sure to identify.
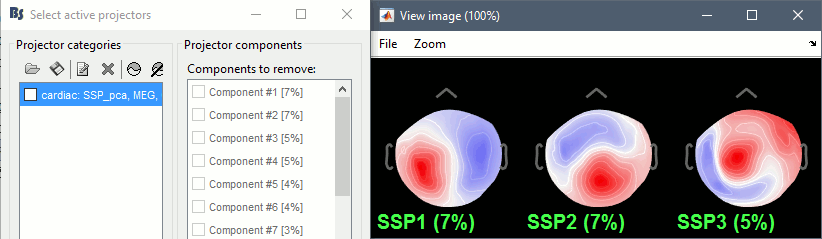
Artifacts > SSP: Eyeblinks: Event "blink", sensors="MEG ", use existing SSP. (select component #1)
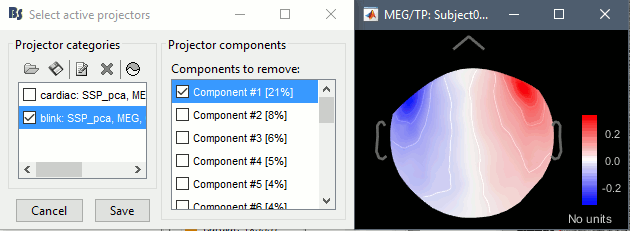
Epoching and averaging
Import the recordings
Right-click on the filtered file "Raw | notch" > Import in dabase.
The following figure appears, and asks how to import these recordings in the Brainstorm database.
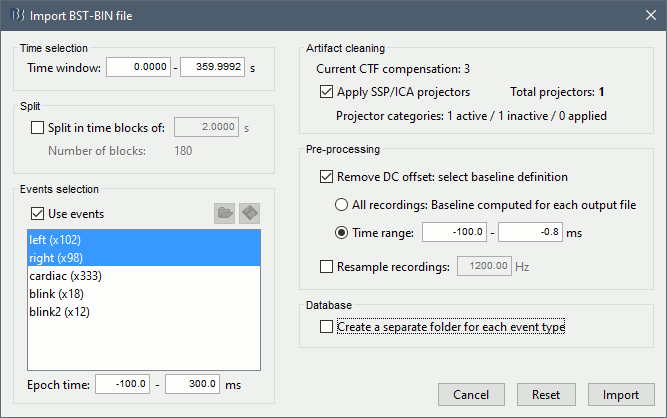
Time window: Time range of interest, keep all the time definition.
Split: Useful to import continuous recordings without events. We do not need this here.
Events selection: Check the "Use events" option, and select both Left and Right.
Epoch time: Time instants that will be extracted before an after each event, to create the epochs that will be saved in the database. Set it to [-100, +300] ms
Use Signal Space Projections: Use the active SSP projectors calculated during the previous pre-processing steps. Keep this option selected.
Remove DC Offset: Check this option, and select: Time range: [-100, 0] ms. For each epoch, this will: compute the average of each channel over the baseline (pre-stimulus interval: -100ms to 0ms), then subtract it from the channel at all the times in [-100,+300]ms.
Resample recordings: Keep this unchecked
Create a separate folder for each epoch type: If selected, a new folder is created for each event type (here, it would create two folders "left" and "right"). This option is mostly for EEG studies with channel files shared across runs. In a MEG study, we usually recommend to use one channel file per run, and to import all the epochs from one run in the same folder.
Click on Import and wait. At the end, you are asked whether you want to ignore one epoch that is shorter than the others. This happens because the acquisition of the MEG signals was stopped less than 300ms after the last stimulus trigger was sent. Therefore, the last epoch cannot have the full [-100,300]ms time definition. This shorter epoch would prevent us from averaging all the trials easily. As we already have enough repetitions in this experiment, we can just ignore it. Answer Yes to this question to discard the last epoch.
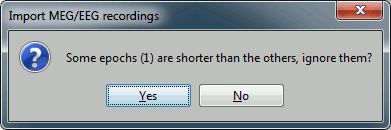
At this stage, you should review all the trials (press F3 to jump to the next file), separately for the magnetometers and the gradiometers, to make sure that you don't have any bad trial that have been imported. If you find a bad trial: right-click on the file or on the figure > Reject trial.
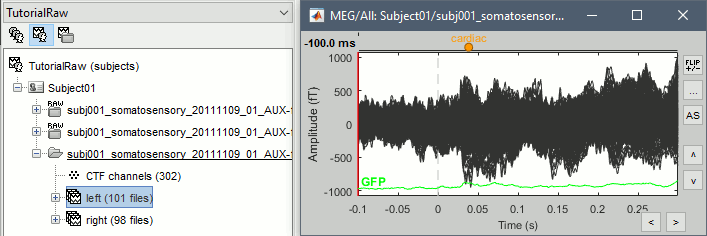
Averaging
- Drag and drop all the left and right trials to the Process1 tab.
Run the process Average > Average files > By trial group (folder average):
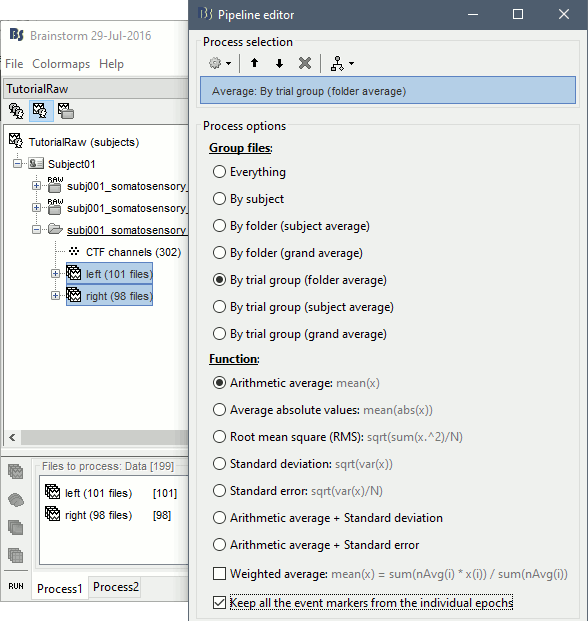
Double-click on the Left and Right averages to display all the MEG sensors:
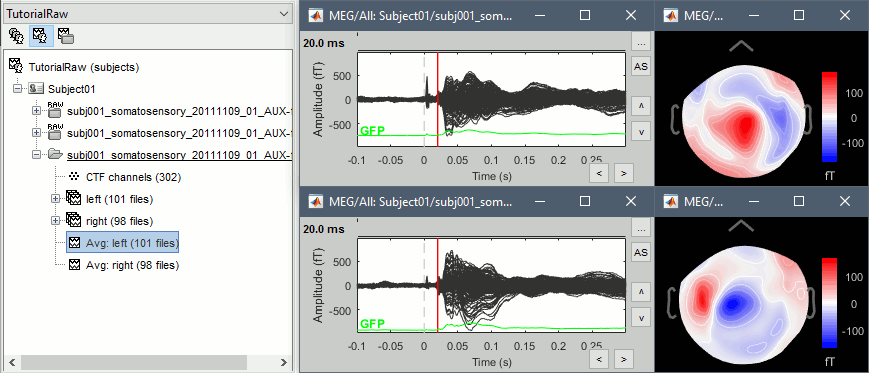
Stimulation artifact
Now zoom around 4ms in time (mouse wheel, or two figures up/down on macbooks) and amplitude (control + zoom). Notice this very strong and sharp peak followed by fast oscillations. This is an artifact due to the electric stimulation device. In the stimulation setup: the stimulus trigger is initiated by the stimulation computer and sent to the electric stimulator. This stimulator generates an electric pulse that is sent to electrodes on the subject's wrists. This electric current flows in the wires and the in the body, so it also generates a small magnetic field that is captured by the MEG sensors. This is what we observe here at 4ms.
This means that whenever we decide to send an electric stimulus, there is a 4ms delay before the stimulus is actually received by the subject, due to all the electronics in between. Which means that everything is shifted by 4ms in the recordings. These hardware delays are unavoidable and should be quantified precisely before starting scanning subjects or patients.
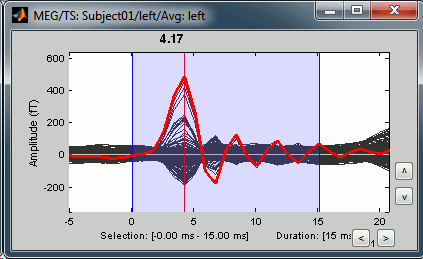
Then you have two options: either you remember it and subtract the delays when you publish your results (there is a risk of forgetting about them), or you fix the files right now by fixing the time reference in all the files (there is a risk of forgetting to fix all the subjects/runs in the same way). Let's illustrate this second method now.
Close all the figures, clear the Process1 list (right-click > clear, or select+delete key), and drag and drop all the trials and all the averages (or simply the two left and right condition folders). Select the process "Pre-process > Add time offset". Set the time offset to -4.2 ms, to bring back this stimulation peak at 0ms. Select also the "Overwrite input files" option, to avoid too many duplications of unnecessary files in the database.
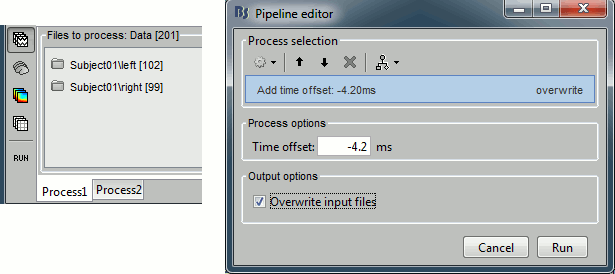
This fixes the individual trials and the averages. Double-click on the "Avg: left" file again to observe that the stimulation artifact is now occurring at 0ms exactly, which means that t=0s represents the time when the electric pulse is received by the subject.
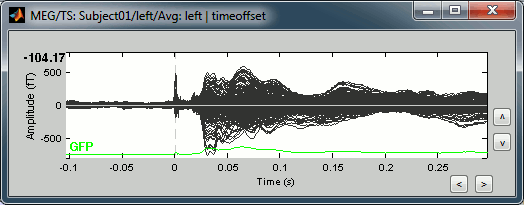
Explore the average
Open the time series for the "Avg: left". Then press Control+T, to see on the side a spatial topography at the current time point. Then observe what is happening between 0ms and 100ms. Start at 0ms and play it like a movie using the arrow keys left and right, to follow the brain activity millisecond by millisecond:
16 ms: First response, the sensory information reaches the right somatosensory cortex (S1)
30 ms: Stronger and more focal activity in the same region, but with a source oriented in the opposite direction (S1)
60 ms: Activity appearing progressively in a more lateral region (S1 + S2)
70 ms: Activity in the same area in the left hemisphere (S2 left + S2 right)
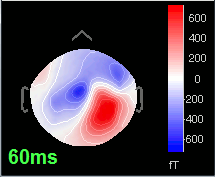
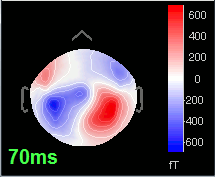
Source analysis
Let's reproduce the same observations at the source level. The concepts related with the source estimation are not discussed here; for more information, refer to the introduction tutorials #6 to #8.
First, delete all the files related with the source estimation calculated in the previous tutorials, available in the (Common files) folder: the head model, the noise covariance and inverse model. We can now provide a better estimate of the noise (affects the inverse model), and we defined new SSP operators (affects the head model).
Head model
Right-click on any node that contains the channel file (including the channel file itself), and select: "Compute head model". Leave all the default options: cortex source space, and overlapping spheres. The lead field matrix is saved in file "Overlapping spheres" in (Common files).
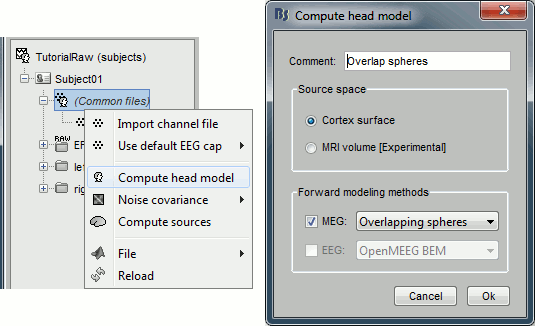
Noise covariance matrix
To estimate the sources properly, we need an estimation of the noise level for each sensor. A good way to do this is to compute the covariance matrix of the concatenation of the baselines from all the trials in both conditions.
- Select at the same time the two groups of trials (right and left). To do this: hold the Control (or Cmd on Macs) key and click successively on the Right and the Left trial lists.
Right-click on one of them and select: Noise covariance > Compute from recordings. Set the baseline to [-104,-5] ms, to consider as noise everything that happens before the beginning of the stimulation artifact. Leave the other options to the default values. Click on Ok.
This operation computes the noise covariance matrix based on the baseline of all the good trials (199 files). The result is stored in a new file "Noise covariance" in the (Common files) folder.
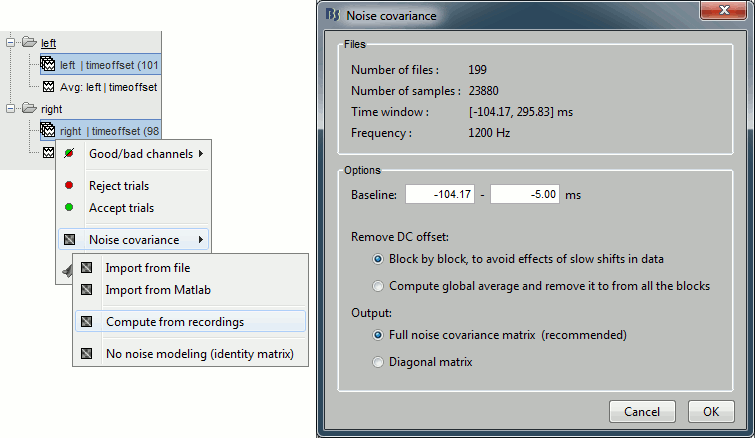
Inverse model
Right-click on (Common files), on the head model or on the subject node, and select "Compute sources". A shared inversion kernel is created in (Common files); a link node is now visible for each recordings file, single trials and averages. For more information about what these links mean and the operations performed to display them, please refer to the tutorial #8 "Source estimation".
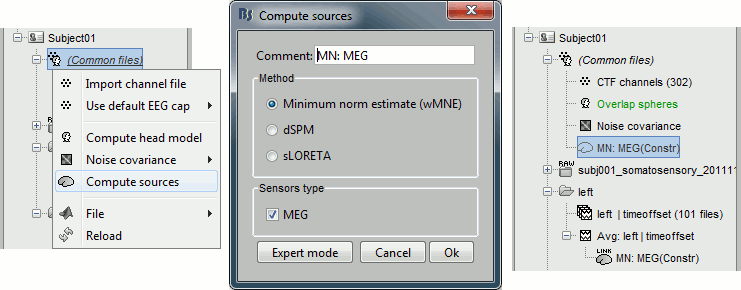
Explore the sources
Right-click on the sources for the left average > Cortical activations > Display on cortex, or simply double click on the file. Go to the main response peak at t = 30ms, and increase the amplitude threshold to 100%. You see a strong activity around the right primary somatosensory cortex, but there are still lots of brain areas that are shown in plain red (value >= 100% maximum of the colorbar ~= 280 pA.m).
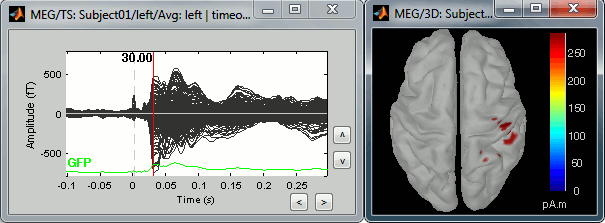
The colorbar maximum is set to an estimation of the maximum source amplitude over the time. This estimation is done by finding the time instant with the highest global field power on the sensors (green trace GFP), estimating the sources for this time only, and then taking the maximum source value at this time point. It is a very fast estimate, but not very reliable; we use it because calculating the full source matrix (all the time points x all the sources) just for finding the maximum value would be too long.
In this case, the real maximum is probably higher than what is used by default. To redefine the colorbar maximum: right-click on the 3D figure > Colormap: sources > Set colorbar max value. Set the maximum to 480 pA.m, or any other value that would lead to see just one very focal point on the brain at 30ms.
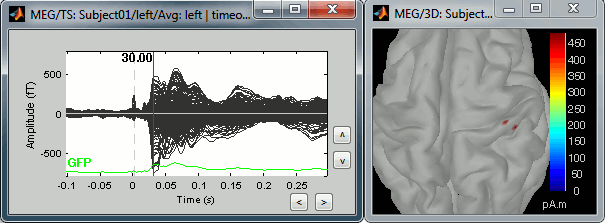
Go back to the first small peak at t=16ms, and lower the threshold to 10%. Then do what you did with at the sensor level: follow the information processing in the brain until 100ms, millisecond by millisecond, adapting the threshold and the camera position when needed:
16 ms (top-left): First response, primary somatosensory cortex (S1 right)
30 ms (top-right): S1 right
60 ms (bottom-left): Secondary somatosensory cortex (S2 right)
70 ms (bottom-right): Activity ipsilateral to the stimulus (S2 left + S2 right)
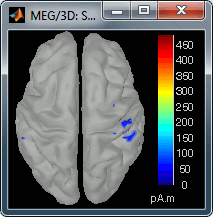
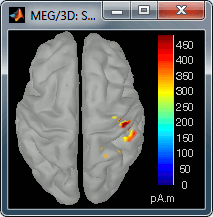
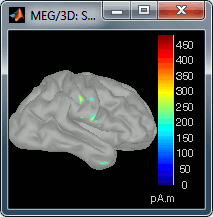
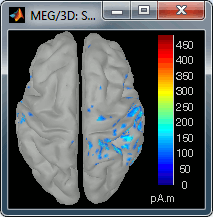
Scripting
The following script from the Brainstorm distribution reproduces the analysis presented in this tutorial page: brainstorm3/toolbox/script/tutorial_raw.m
