|
Size: 4062
Comment:
|
Size: 6643
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 4: | Line 4: |
| = Tutorial: Import and visualize functional NIRS data = | |
| Line 16: | Line 17: |
| = Download = | == Download == |
| Line 22: | Line 23: |
| = Presentation of the experiment = * One subject, two acquisition runs of /!\ ''' XXX ''' minutes * Finger tapping task: /!\ ''' XXX ''' stimulation blocks of 30 seconds each, inter-stimulus interval of /!\ ''' XXX ''' seconds * /!\ ''' XXX ''' sources and /!\ ''' XXX ''' detectors arranged in a /!\ ''' XXX ''' montage, with /!\ ''' XXX ''' proximity channels * Two wavelengths: 690nm and 860nm |
== Presentation of the experiment == * One subject, one acquisition run of /!\ ''' XXX ''' minutes * Finger tapping task: 10 stimulation blocks of 30 seconds each, inter-stimulus interval of /!\ ''' XXX ''' seconds * 4 sources and 12 detectors (+ 4 proximity channels) placed above the right motor region * Two wavelengths: 690nm and 830nm |
| Line 29: | Line 30: |
| = Create Data structure = | == Create the data structure == Create a protocol called "TutorialNIRSTORM": |
| Line 31: | Line 33: |
| Create a protocol called "TutorialNIRSTORM": * Got to File -> New Protocol * Use the following setting : * '''Default anatomy''': Use individual anatomy. * '''Default channel file''': Use one channel file per subject (EEG). In term of sensor configuration, NIRS is very similar to EEG and the placement of optodes may change from subject to the other. (!) Should we add (EEG '''or NIRS''') in the interface? |
* Got to File -> New Protocol * Use the following setting : * '''Default anatomy''': Use individual anatomy. * '''Default channel file''': Use one channel file per subject (EEG). In term of sensor configuration, NIRS is very similar to EEG and the placement of optodes may change from subject to the other. (!) Should we add (EEG '''or NIRS''') in the interface? |
| Line 43: | Line 42: |
| = Import anatomy = | == Import anatomy == === Import MRI === Make sure you are in the anatomy view of the protocol. Right-click on "Subject01 -> Import MRI". Select T1_MRI.nii from the NIRS_sample data folder. This will open the MRI review panel where you have to set the fudicial points (See [[http://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy|Import the subject anatomy]]). |
| Line 45: | Line 46: |
| == Import MRI == Make sure you are in the anatomy view of the protocol. Right-click on "Subject01 -> Import MRI". Select T1_MRI.nii from the NIRS_sample data folder. This will open the MRI review panel where you have to set the fudicial points (See [[http://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy|Import the subject anatomy]]). == Import Meshes == |
=== Import Meshes === |
| Line 59: | Line 53: |
| /!\ '''Add screenshot''' | {{attachment:NIRSTORM_tut1_new_MRI_meshes.gif||height="335",width="355"}} == Import NIRS functional data == The functional data used in this tutorial was exported from the Brainsight acquisition software and is available in the NIRS sample folder in S01_Block_FO_LH_Run01.bs. This folder contains the following files: * fiducials.txt: the coordinates of the fudicials (nasion, left ear, right ear).<<BR>>These positions should have been digitized at the same location as the fiducials previously marked on the anatomical MRI. These points will be used by Brainstorm for the registration, hence the consistency between the digitized and marked fiducials is essential for good * optodes.txt: the coordinates of the optodes (sources and detectors), in the same referential as for the fiducials. Note: the actual referential is not relevant here, as the registration will be performed by Brainstorm afterwards. * S01_Block_FO_LH_Run01.nirs: the NIRS data in a HOMer-based format /!\ '''document format'''.<<BR>>Note: The fields ''SrcPos'' and ''DetPos'' will be overwritten to match the given coordinates in "optodes.txt" To import this data set in Brainstorm: * Go to the "functional data" view of the protocol. * Right-click on "Subject01 -> Import MEG/EEG/NIRS" * Select file type "NIRS: BS (.bs)" |
| Line 62: | Line 69: |
| import Brainvisa head mesh and MRI file + provide datasets | |
| Line 64: | Line 70: |
| mark fudicials | * Load the folder "S01_Block_FO_LH_Run01.bs" in the NIRS sample folder. * Refine registration now? '''YES'''<<BR>>This operation is detailed in the next section |
| Line 66: | Line 73: |
| = Import NIRS functional data = | == Registration == As in the tutorial The resitration between the MRI and the NIRS is done in two steps |
| Line 68: | Line 76: |
| Go to the "functional data" view of the protocol. | == Review Channel information == The resulting data organization should be: |
| Line 70: | Line 79: |
| NIRS Acquisition + description | {{attachment:func_data_loading_result.gif}} |
| Line 72: | Line 81: |
| dataset files: | This indicates that the data comes from the Brainsight system (BS) and comprises 97 channels. |
| Line 74: | Line 83: |
| * 2 nirs files with proper SrcPos and DetPos fields | To review the content of channels, right-click on "BS channels -> Edit channel file". || ||'''Name''' ||'''Type''' ||'''Group''' ||'''Comment''' ||'''Loc(1)''' ||'''Loc(2)''' ||'''Loc(3)''' ||... || ||1 ||TAPPING ||Stim || || ||N/A ||N/A ||N/A || || ||2 ||WLs ||NIRS_WL_DEF || || ||N/A ||N/A ||N/A || || ||3 ||S1D1WL1 ||NIRS_WL1 || || ||coords S1 ||coords D1 ||coords middle [S1-D1] || || ||4 ||S1D1WL2 ||NIRS_WL2 || || ||coords S1 ||coords D1 ||coords middle [S1-D1] || || ||5 ||S1D2WL1 ||NIRS_WL1 || || ||coords S1 ||coords D2 ||coords middle [S1-D2] || || ||6 ||S1D2WL2 ||NIRS_WL2 || || ||coords S1 ||coords D2 ||coords middle [S1-D2] || || ||7 ||S1P1WL1 ||NIRS_WL1 ||NIRS_PROX || ||coords S1 ||coords D2 ||coords middle [S1-D2] || || ||8 ||S1P1WL2 ||NIRS_WL2 ||NIRS_PROX || ||coords S1 ||coords D2 ||coords middle [S1-D2] || || |
| Line 76: | Line 94: |
| -> generate these from brainsight output merged with montage coords | |
| Line 78: | Line 95: |
| -> check if there is a way to directly have this from brainsight without having to forge it | |
| Line 80: | Line 96: |
| * 1 file of digitized coords of fiducials (NAS, LE, RE) as a txt file. Coords should be in the same referential as SrcPos and DetPos | * The channel named "TAPPING" encodes the stimulation paradigm * The channel named "WLs" defines the set of wavelengths * Other channels contain the NIRS time-series measurements. For a given NIRS channel, its name is composed of the pair Source / Detector and the wavelength index. Column Loc(1) contains the coordinates of the source, Loc(2) the coordinates of the associated detector and Loc(3) the coordinates of the middle point between the source and the detector. * The group "NIRS_PROX" indicates that the channel is a close-source measurement. |
|
|
Tutorial: Import and visualize functional NIRS data
Authors: Thomas Vincent, Zhengchen Cai
The current tutorial assumes that the tutorials 1 to 5 have been performed. Even if they focus on MEG data, they introduce Brainstorm features that are used in this tutorial.
List of prerequisites:
Download
The dataset used in this tutorial is available online .
Go to the Download
 link page of this website, and download the file: nirs_sample.zip
link page of this website, and download the file: nirs_sample.zip - Unzip it in a folder that is not in any of the Brainstorm folders
Presentation of the experiment
One subject, one acquisition run of
 XXX minutes
XXX minutes Finger tapping task: 10 stimulation blocks of 30 seconds each, inter-stimulus interval of
 XXX seconds
XXX seconds - 4 sources and 12 detectors (+ 4 proximity channels) placed above the right motor region
- Two wavelengths: 690nm and 830nm
MRI anatomy 3T from
 scanner type
scanner type
Create the data structure
Create a protocol called "TutorialNIRSTORM":
Got to File -> New Protocol
- Use the following setting :
Default anatomy: Use individual anatomy.
Default channel file: Use one channel file per subject (EEG).
In term of sensor configuration, NIRS is very similar to EEG and the placement of optodes may change from subject to the other. ![]() Should we add (EEG or NIRS) in the interface?
Should we add (EEG or NIRS) in the interface?
Create a subject called "Subject01" (Go to File -> New subject), with the default options
Import anatomy
Import MRI
Make sure you are in the anatomy view of the protocol. Right-click on "Subject01 -> Import MRI". Select T1_MRI.nii from the NIRS_sample data folder. This will open the MRI review panel where you have to set the fudicial points (See Import the subject anatomy).
Import Meshes
The head and white segmentations provided in the NIRS sample data were computed with Brainvisa.
Right-click on "Subject01 -> Import surfaces". From the NIRS sample data folder, select files: head_10000V.mesh, hemi_8003V.mesh and white_8003V.mesh.
You can check the regristration between the MRI and the loaded meshes by right-clicking on each meash element and going to "MRI registration -> Check MRI/Surface registration".
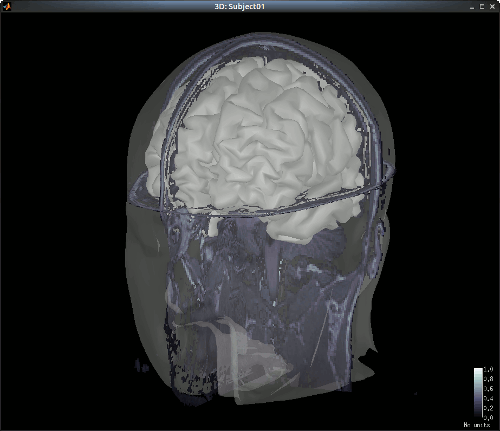
Import NIRS functional data
The functional data used in this tutorial was exported from the Brainsight acquisition software and is available in the NIRS sample folder in S01_Block_FO_LH_Run01.bs. This folder contains the following files:
fiducials.txt: the coordinates of the fudicials (nasion, left ear, right ear).
These positions should have been digitized at the same location as the fiducials previously marked on the anatomical MRI. These points will be used by Brainstorm for the registration, hence the consistency between the digitized and marked fiducials is essential for good- optodes.txt: the coordinates of the optodes (sources and detectors), in the same referential as for the fiducials. Note: the actual referential is not relevant here, as the registration will be performed by Brainstorm afterwards.
S01_Block_FO_LH_Run01.nirs: the NIRS data in a HOMer-based format
 document format.
document format.
Note: The fields SrcPos and DetPos will be overwritten to match the given coordinates in "optodes.txt"
To import this data set in Brainstorm:
- Go to the "functional data" view of the protocol.
Right-click on "Subject01 -> Import MEG/EEG/NIRS"
- Select file type "NIRS: BS (.bs)"
- Load the folder "S01_Block_FO_LH_Run01.bs" in the NIRS sample folder.
Refine registration now? YES
This operation is detailed in the next section
Registration
As in the tutorial The resitration between the MRI and the NIRS is done in two steps
Review Channel information
The resulting data organization should be:
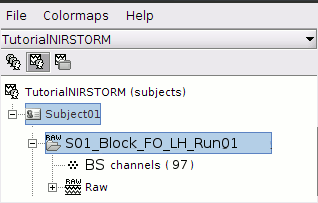
This indicates that the data comes from the Brainsight system (BS) and comprises 97 channels.
To review the content of channels, right-click on "BS channels -> Edit channel file".
|
Name |
Type |
Group |
Comment |
Loc(1) |
Loc(2) |
Loc(3) |
... |
1 |
TAPPING |
Stim |
|
|
N/A |
N/A |
N/A |
|
2 |
WLs |
NIRS_WL_DEF |
|
|
N/A |
N/A |
N/A |
|
3 |
S1D1WL1 |
NIRS_WL1 |
|
|
coords S1 |
coords D1 |
coords middle [S1-D1] |
|
4 |
S1D1WL2 |
NIRS_WL2 |
|
|
coords S1 |
coords D1 |
coords middle [S1-D1] |
|
5 |
S1D2WL1 |
NIRS_WL1 |
|
|
coords S1 |
coords D2 |
coords middle [S1-D2] |
|
6 |
S1D2WL2 |
NIRS_WL2 |
|
|
coords S1 |
coords D2 |
coords middle [S1-D2] |
|
7 |
S1P1WL1 |
NIRS_WL1 |
NIRS_PROX |
|
coords S1 |
coords D2 |
coords middle [S1-D2] |
|
8 |
S1P1WL2 |
NIRS_WL2 |
NIRS_PROX |
|
coords S1 |
coords D2 |
coords middle [S1-D2] |
|
- The channel named "TAPPING" encodes the stimulation paradigm
- The channel named "WLs" defines the set of wavelengths
- Other channels contain the NIRS time-series measurements. For a given NIRS channel, its name is composed of the pair Source / Detector and the wavelength index. Column Loc(1) contains the coordinates of the source, Loc(2) the coordinates of the associated detector and Loc(3) the coordinates of the middle point between the source and the detector.
- The group "NIRS_PROX" indicates that the channel is a close-source measurement.
NIRS-MRI coregistration
Use automatic registration
Keep only step 1
Display Optodes (adapted from “Display Sensors”)
- no helmet here -> should be able to show optode positions over head meshSKip manual registration
Edit the channel file
Introduce new nomenclature
S1D1WL1 NIRS_WL1 …S1D1WL2 NIRS_WL2 …
Visualize NIRS signals
Depends on tut #5
Introduce default channel groups: ALL, WL1, WL2
Just adapt sections “Montage selection” and “Channel selection”
