|
Size: 7603
Comment:
|
Size: 7598
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 110: | Line 110: |
| {{{#!wiki information | {{{#!wiki attention |
| Line 112: | Line 112: |
| To facilitate trial averaging, it is recommended that you duplicate the events linked to your task and convert them to simple events using the start of each trial. For this tutorial, duplicate the event 'tapping', convert it to simple, and rename it tapping/start. | To facilitate trial averaging, it is recommended that you duplicate the events linked to your task and convert them to simple events using the start of each trial. For this tutorial, duplicate the event 'tapping', convert it to simple, and rename it tapping/start. |
| Line 114: | Line 114: |
NIRSTORM: a Brainstorm plugin specialized in the analysis of fNIRS data

Author: Édouard Delaire, Lea Larreur, Dr. Zhengchen Cai, Dr. Christophe Grova
This tutorial aims to introduce NIRSTORM. NISTORM is a plug-in dedicated to the analysis of functional near-infrared spectroscopy (fNIRS) data inside Brainstorm.
This tutorial will cover three main aspects of NIRSTORM:
- Data importation and preprocessing
- Data analysis at the channel level
- Data analysis on the cortical surface.
This tutorial assumes that you are already familiar with the Brainstorm environment. If you are not, we encourage you to follow the tutorial 1 to 23.
Contents
Introduction
Presentation of the experiment
The data presented in this tutorial correspond to a finger tapping task perform by one subject using their left, non-dominant hand. NIRS montage was done to target the subject right-motor cortex.
Data description:
• One subject, one run of 19 minutes acquired with a sampling rate of 10Hz on a CW fNIRS Brainsight device (Rogue-Research Inc., Montreal, Canada)
• Finger tapping task: 20 blocks of 10 seconds each, with a rest period of 30 to 60 seconds.
• 3 sources, 15 detectors, and 1 proximity detector.
• 2 wavelengths: 685nm and 830nm
• 3T MRI anatomy processed by FreeSurfer 5.3.0.
Download nirstorm_tutorial_2024.zip from https://osf.io/md54y/?view_only=0d8ad17d1e1449b5ad36864eeb3424ed
Setup
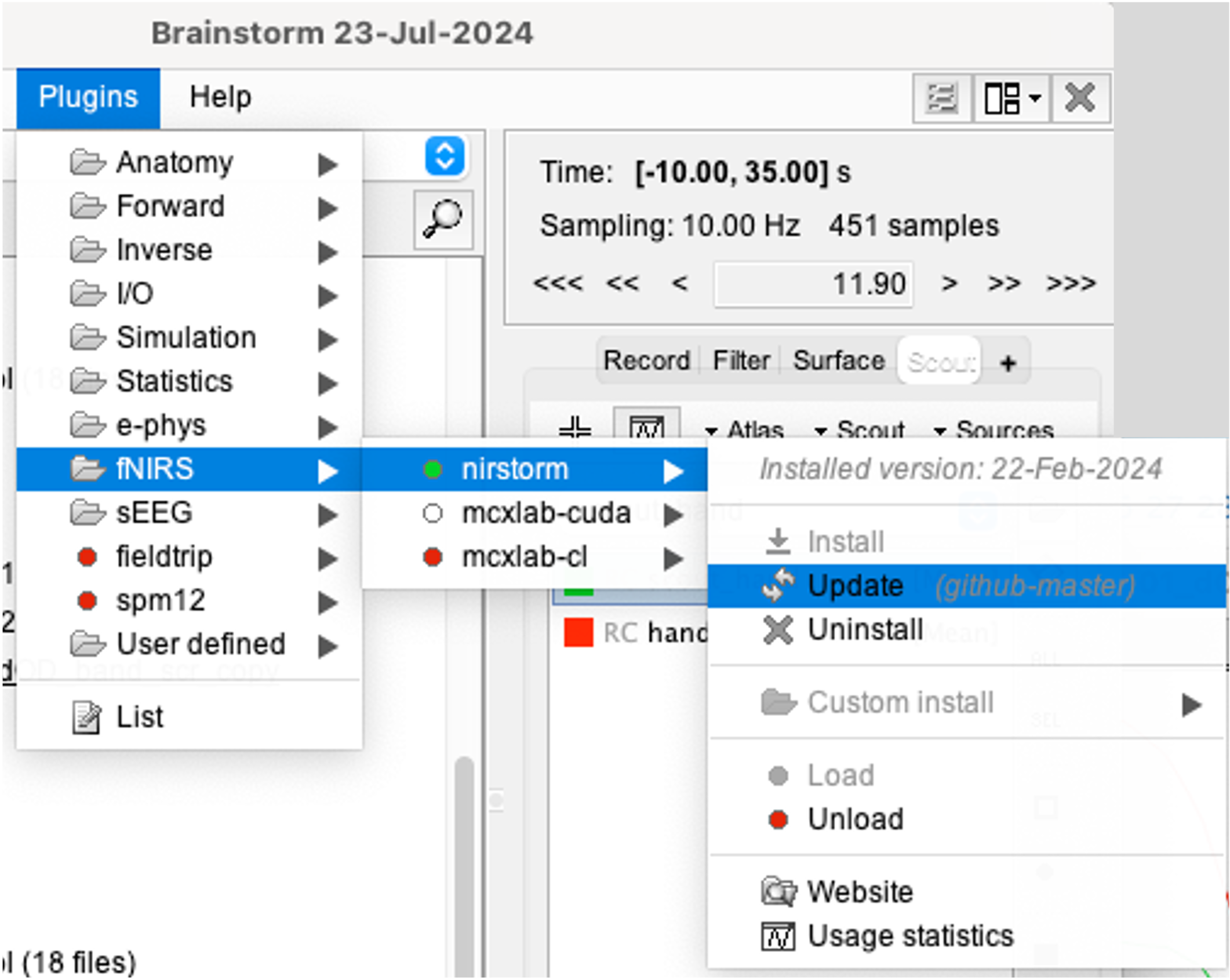
You can install NIRSTORM manually from Brainstorm directly. For that, go to Plug-ins > fNIRS > nirstorm.
Data Importation
- Start Brainstorm (Matlab scripts or stand-alone version)
Select the menu File > Create new protocol. Name it "TutorialNIRS" and select the options:
"No, use individual anatomy",
"No, use one channel file per acquisition run (MEG/EEG)".
In term of sensor configuration, NIRS is similar to EEG and the placement of optodes may change between subjects. Also, the channel definition will change during data processing, that's why you should always use one channel file per acquisition run, even if the optode placement does not change.
Import the anatomy
- Switch to the "anatomy" view of the protocol.
Right-click on the TutorialNIRS folder > New subject > sub-01
- Leave the default options you set for the protocol
Right-click on the subject node > Import anatomy folder:
- Set the file format: "Freesurfer + volumes atlases"
Select the folder: nirstorm_tutorial_2024/derivatives/FreeSurfer/sub-01
- Number of vertices of the cortex surface: 15000 (default value)
- Answer "yes" when asked to apply the transformation.
Set the 3 required fiducial points, indicated below in (x,y,z) MRI coordinates. You can right-click on the MRI viewer > Edit fiducial positions > MRI coordinates, and copy-paste the following coordinates in the corresponding fields. Click [Save] when done.
- NAS: 131, 213, 117
- LPA: 51, 116, 80
- RPA: 208, 117, 86
- AC: 131, 128, 130
- PC: 129, 103, 124
- IH: 133, 135, 176
For more information about importing subject anatomy, consult: https://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy
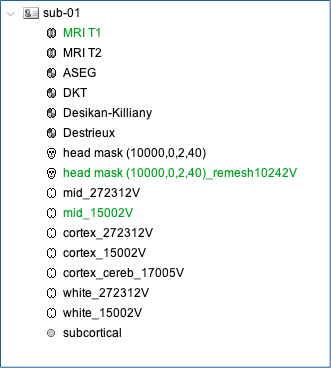
- Once the anatomy is imported, make sure that the surface "mid_15002V" is selected (green). If it is not, right-click on it and select "set as default cortex".
- Resample the head mask to get a uniform mesh on the head. For that, right click on head mask and select remesh, then 10242 vertices
At this point, your database should looks like this:
Review the nirs functional data
The functional data used in this tutorial was produced by the Brainsight acquisition software and is available in the data subfolder of the nirs sample folder(nirstorm_tutorial_2024/sub-01/nirs). The data were exported according to the BIDS format:
See https://bids-specification.readthedocs.io/en/stable/modality-specific-files/near-infrared-spectroscopy.html for a description of each files.
To import this dataset in Brainstorm:
- Go to the "functional data" view of the protocol.
Right-click on Subject01 > Review raw file
Select file type NIRS: SNIRF (.snirf)
- Select file nirstorm_tutorial_2024/sub-01/nirs/sub-01_task-tapping_run-01.snirf
Registration
In the same way as in the tutorial "Channel file / MEG-MRI coregistration", the registration between the MRI and the NIRS is first based on three reference points Nasion, Left and Right ears. It can then be refined with the either the full head shape of the subject or with manual adjustment.
The initial registration is based on the three fiducial point that define the Subject Coordinate System (SCS): nasion, left ear, right ear. You have marked these three points in the MRI viewer in the previous part.
- These same three points have also been marked before the acquisition of the NIRS recordings. The person who recorded this subject digitized their positions with a tracking device (here Brainsight). The position of these points are saved in the NIRS datasets (see fiducials.txt).
- When the NIRS recordings are loaded into the Brainstorm database, they are aligned on the MRI using these fiducial points: the NAS/LPA/RPA points digitized with Brainsight are matched with the ones we placed in the MRI Viewer.
To review this registration:
Right-click on NIRS-BRS sensors (97) > Display sensors > NIRS (pairs).This will display sources as red balls and detectors as green balls. Source/detector pairings are displayed as blue lines.
Right-click on NIRS-BRS sensors (97) > Display sensors > NIRS (scalp).This will display sources as red balls and detectors as green balls
To check the coregistration, Right-click on NIRS-BRS sensors (97) > MRI registration > check.
For more information about the coregistration, consult "Channel file / MEG-MRI coregistration"
Review of recording
Select "sub-01 |- sub-01_task-tapping_run-01 |- Link to raw file -> NIRS -> Display time series". It will open a new figure with superimposed channels.
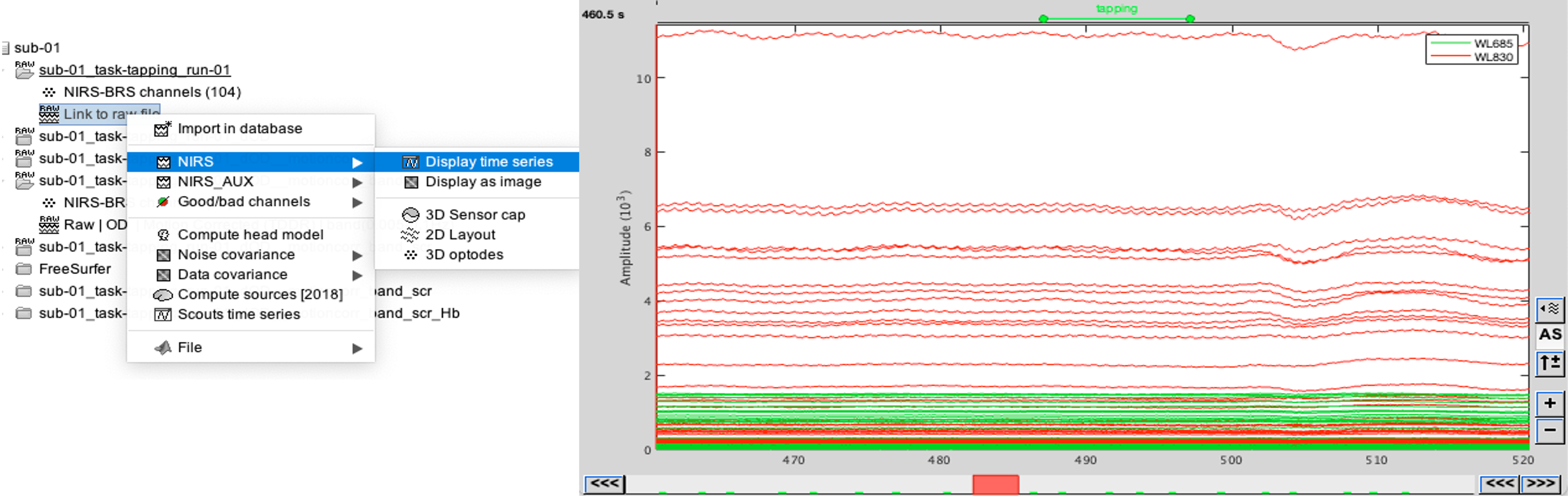
If coloring is not visible, right-click on the figure the select "Montage > NIRS Overlay > NIRS Overlay" Indeed, brainstorm uses a dynamical montage, called NIRS Overlay, to regroup and color-code nirs time-series depending on the wavelength (red: 830nm, green:686nm). The signals for a given pair of source and detectors are also grouped when using the selection tool. So clicking one curve for one wavelength will also select the other wavelength for the same pair.
To import the events corresponding to your paradign, consult https://neuroimage.usc.edu/brainstorm/Tutorials/EventMarkers
To facilitate trial averaging, it is recommended that you duplicate the events linked to your task and convert them to simple events using the start of each trial. For this tutorial, duplicate the event 'tapping', convert it to simple, and rename it tapping/start.
Preprocessing
