|
Size: 6246
Comment:
|
Size: 6147
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 9: | Line 9: |
| When using a source model with unconstrained orientations, i.e. 3 components (x,y,z) at each location, one may wish to reduce the results to a single source orientation per location, a "flat" map, for some analyses. In many cases, e.g. spectral or connectivity measures, the signal phase must be preserved, which precludes taking the norm of the 3 components. In such cases, a single orientation can be selected with Principal Component Analysis (PCA), such that the resulting time series has maximal power, i.e. the largest kept variance from the initial 3 components. | Further analyses of source models with unconstrained orientations, i.e. with 3 components (x,y,z) at each source location, are often more convenient after a "flat" source is obtained, whereby the source time series are reduced to one single dimension per source location, as with orientation-constrained source maps. Importantly and in many cases, e.g., spectral or connectivity analyses, the phase of the time series must be preserved. Reducing the 3 local dimensions to the norm of the 3 components introduces nonlinear distorsions and must be avoided. A single equivalent source orientation can be estimated with Principal Component Analysis (PCA); the PCA approach yields a resulting summary time series with maximum variance (equivalent to signal power). |
| Line 11: | Line 11: |
| This can be done directly with the process "'''Sources > Unconstrained to flat map'''", which will save a flattened copy of source maps (full or kernel) in the Brainstorm tree, or optionally through other processes such as connectivity, where the flat maps are saved to temporary files and don't appear in the tree. | Such flattening can be derived with the process "'''Sources > Unconstrained to flat map'''", which saves a flattened copy of source maps (full or kernel) in the Brainstorm data tree, or optionally directly through other processes such as connectivity, where the flat maps are saved to temporary files and are not saved in the tree. |
| Line 13: | Line 13: |
| == Sign of principal component == Since PCA finds a combination of sources that maximizes power, i.e. the sum of the signal squared, the resulting sign of the component is arbitrary. Brainstorm attempts to pick a sign that will best reflect the original data and, for scouts, the cortex geometry. However, prior to 2023 the procedure was not robust and resulted in arbitrary signs. This can be an issue for subsequent analysis steps, e.g. leading to cancellations when averaging across epochs or files, or for some connectivity measures. Brainstorm now offers 2 options that properly deal with the component sign across epochs and conditions as explained in the next section, but this requires that all files are processed together. |
== An imporant detail: consistency of the signal polarity of the principal component == The polarity of the PCA component is arbitrary in the sense that it is driven by a mathematical factorisation of the original time series, not by physiology. In June 2023, we have improved the Brainstorm's approach to assign a consistent polarity of the PCA time series to best reflect the original data and, for scouts, the cortex geometry. Brainstorm features 2 options in that purpose as explained in the next section. |
| Line 16: | Line 16: |
| Note that the sign issue just discussed is independent from the ambiguity on the sign of constrained sources due to the geometry of the cortex and resolution of the inverse model, as explained in the [[https://neuroimage.usc.edu/brainstorm/Tutorials/Scouts#Sign_flip|Scout tutorial]]. That sign flipping of individual sources is used for the 'mean' scout function, but this is automatically dealt with when using PCA, while it is the overall sign of the resulting component that remains ambiguous. | Note that this aspect of polarity assignment is independent from the inherent ambiguity on the polarity of MEG/EEG source time series, as explained in the [[https://neuroimage.usc.edu/brainstorm/Tutorials/Scouts#Sign_flip|Scout tutorial]]. This means that even though Brainstorm's processes maximizes the consistency of the polarity of source time series across scouts, trials and other dimensions of a study, the resulting polarity remains ambiguous. |
| Line 18: | Line 18: |
| Note that sign ambiguity (from both these issues) is still present at the subject level and appropriate steps should be taken for group analysis (e.g. rectify). The issue can also be present within subject if files or conditions are processed independently. | In particular, signal polarity might vary across participants and therefore group analyses may require to pool rectified (polarity-free) versions of source signals, to maximize the detection of effects across participants. |
Flattening of orientation-unconstrained source models
Author: Marc Lalancette
This section details how Principal Component Analysis (PCA) is used in Brainstorm for dimension reduction, in particular for source models with unconstrained orientations.
Contents
Flattening of source maps using principal component analysis
Further analyses of source models with unconstrained orientations, i.e. with 3 components (x,y,z) at each source location, are often more convenient after a "flat" source is obtained, whereby the source time series are reduced to one single dimension per source location, as with orientation-constrained source maps. Importantly and in many cases, e.g., spectral or connectivity analyses, the phase of the time series must be preserved. Reducing the 3 local dimensions to the norm of the 3 components introduces nonlinear distorsions and must be avoided. A single equivalent source orientation can be estimated with Principal Component Analysis (PCA); the PCA approach yields a resulting summary time series with maximum variance (equivalent to signal power).
Such flattening can be derived with the process "Sources > Unconstrained to flat map", which saves a flattened copy of source maps (full or kernel) in the Brainstorm data tree, or optionally directly through other processes such as connectivity, where the flat maps are saved to temporary files and are not saved in the tree.
An imporant detail: consistency of the signal polarity of the principal component
The polarity of the PCA component is arbitrary in the sense that it is driven by a mathematical factorisation of the original time series, not by physiology. In June 2023, we have improved the Brainstorm's approach to assign a consistent polarity of the PCA time series to best reflect the original data and, for scouts, the cortex geometry. Brainstorm features 2 options in that purpose as explained in the next section.
Note that this aspect of polarity assignment is independent from the inherent ambiguity on the polarity of MEG/EEG source time series, as explained in the Scout tutorial. This means that even though Brainstorm's processes maximizes the consistency of the polarity of source time series across scouts, trials and other dimensions of a study, the resulting polarity remains ambiguous.
In particular, signal polarity might vary across participants and therefore group analyses may require to pool rectified (polarity-free) versions of source signals, to maximize the detection of effects across participants.
PCA options
PCA is used in various processes within Brainstorm to combine multiple signals, such as sources in a scout or sensors in a cluster, and the options described here apply to all uses of PCA.
PCA now works best when processing all files from a subject at once, and therefore it is important to provide all the epochs and conditions together when running any process where a PCA option is selected.
Each process that offers PCA as an option also includes an "Edit" button to specify further options specific to the PCA computation:
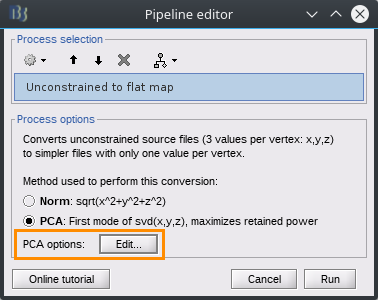
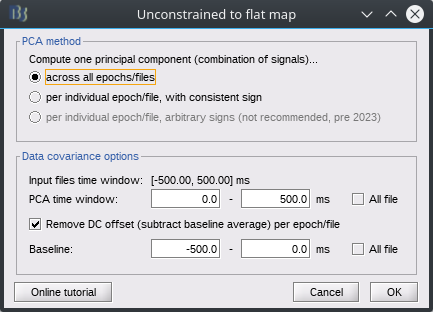
PCA method: affects how different files (epochs, conditions) are treated, and in particular the potential sign issue described above.
across all epochs/files: This results in the same component being applied to each file. It is generally recommended as results are then simpler to interpret, especially for within subject comparisons, e.g. differences across runs or conditions. It can save shared kernels when flattening, and runs faster with kernel link source files.
per individual epoch/file, with consistent sign: A different component is computed for each file, but the sign is corrected to best align with a reference component, obtained with the "across epochs" method. Can be useful for single-trial analysis, while still allowing combining epochs. Slower. Saves individual files.
per individual epoch/file, arbitrary signs: Method used prior to June 2023, no longer recommended due to sign inconsistency. Can be used for single files.
Data covariance options: These options affect only the computation of the principal component coefficients ("loadings", weights used to combine the signals), which are then applied to the unmodified data (without offset removal). These are the same options as when computing the Data Covariance for an adaptive inverse source model. Note that when selecting the 3rd PCA method, these options are fixed to reproduce the behavior prior to June 2023: full time window for baseline and data.
Scaling
A PCA time series represents a fraction of the total activity among all signals, and therefore has a larger amplitude than the average activity across those same signals, and usually scales with the number of signals, e.g. the size of a scout. While the absolute scale of a component may not be very meaningful, Brainstorm now rescales the amplitude of scout PCA time series to be less dependent on the number of vertices in the scout, and to be more comparable with the result of averaging.
A PCA component combines all signals with coefficients constrained such that they have a vector norm of 1, i.e. sum of square coefs = 1. This contrasts with averaging, where each coefficient is 1/N, and their vector norm is sqrt(N * 1/N^2) = sqrt(1/N). Thus scout PCA components are now multiplied by sqrt(1/N).
History
Files resulting from a PCA decomposition store the "% of signal power kept" by the first principal component in the file History, which can be viewed through the tree context menu > File > View file history. (These messages were previously displayed in the command window.)
The history also stores the list of files used to compute the decomposition for PCA "across all epochs/files".
