|
Size: 6328
Comment:
|
Size: 6313
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 7: | Line 7: |
| This tutorial describes how to process and analyze Positron Emission Tomography ([[https://en.wikipedia.org/wiki/Positron_emission_tomography|PET]]) data within Brainstorm and how to extend PET data to surface-based, multimodal analysis. It guides users through importing and pre-processing PET volumes and outlines steps for surface-based analysis of cortical tracer uptake values. PET is a powerful imaging technique widely used in medical and scientific research to study metabolic and functional processes in the brain and body. PET processing involves a series of computational and analytical steps to transform raw data into meaningful visualizations and quantitative insights. These steps ensure accurate reconstruction, correction, and analysis of PET scans, enabling researchers and clinicians to draw precise conclusions. The increasing availability of multimodal datasets that combine PET, MRI and M/EEG provide unique opportunities for integrated analysis of neurophysiology, brain structure and molecular processses mapped by PET radioligands. | This tutorial describes how to process and analyze Positron Emission Tomography ([[https://en.wikipedia.org/wiki/Positron_emission_tomography|PET]]) data within Brainstorm and how to extend PET data to surface-based, multimodal analysis. It guides users through importing and pre-processing PET volumes and outlines steps for surface-based analysis of cortical tracer uptake values. PET is a powerful imaging technique widely used in medical and scientific research to study metabolic and functional processes in the brain and body. PET processing involves a series of computational and analytical steps to transform raw data into meaningful visualizations and quantitative insights. These steps ensure accurate reconstruction, correction, and analysis of PET scans, enabling researchers and clinicians to draw precise conclusions. The increasing availability of multimodal datasets that combine PET, MRI and M/EEG provide unique opportunities for integrated analysis of neurophysiology, brain structure and molecular processes mapped by PET radioligands. |
| Line 10: | Line 10: |
<<BR>> |
|
| Line 77: | Line 75: |
PET processing in Brainstorm
Authors: Diellor Basha
Introduction
This tutorial describes how to process and analyze Positron Emission Tomography (PET) data within Brainstorm and how to extend PET data to surface-based, multimodal analysis. It guides users through importing and pre-processing PET volumes and outlines steps for surface-based analysis of cortical tracer uptake values. PET is a powerful imaging technique widely used in medical and scientific research to study metabolic and functional processes in the brain and body. PET processing involves a series of computational and analytical steps to transform raw data into meaningful visualizations and quantitative insights. These steps ensure accurate reconstruction, correction, and analysis of PET scans, enabling researchers and clinicians to draw precise conclusions. The increasing availability of multimodal datasets that combine PET, MRI and M/EEG provide unique opportunities for integrated analysis of neurophysiology, brain structure and molecular processes mapped by PET radioligands.
This page provides an overview of the essential methods, tools, and considerations involved in PET processing, aiming to support both newcomers and experienced users of Brainstorm. If you are new to Brainstorm, refer to the comprehensive introductroy materials in Brainstorm introduction tutorials to familiarize yourself with the layout and workflow in Brainstorm.
Method Overview
The Brainstorm extension for PET supports importing, processing, registering, visualizing, and analyzing PET data within Brainstorm. PET functionality and workflow are designed to facilitate the analysis of multimodal neuroimaging data by allowing the user to co-analyze MEG, PET and MRI-derived data. Multi-frame (4D) PET volumes are realigned during import and aggregated over time from the 4D image (i.e. mean of the n frames) to obtai a unique 3D volume for co-registration with the subject’s structural MRI. Multi-frame PET volumes are realigned using SPM’s realign and reslice functions and co-registered with SPM. The aggregated and co-registered 3D volume is masked and rescaled to obtain voxel-wise standardized uptake value ratio (SUVR) which are then projected to the cortical surface.
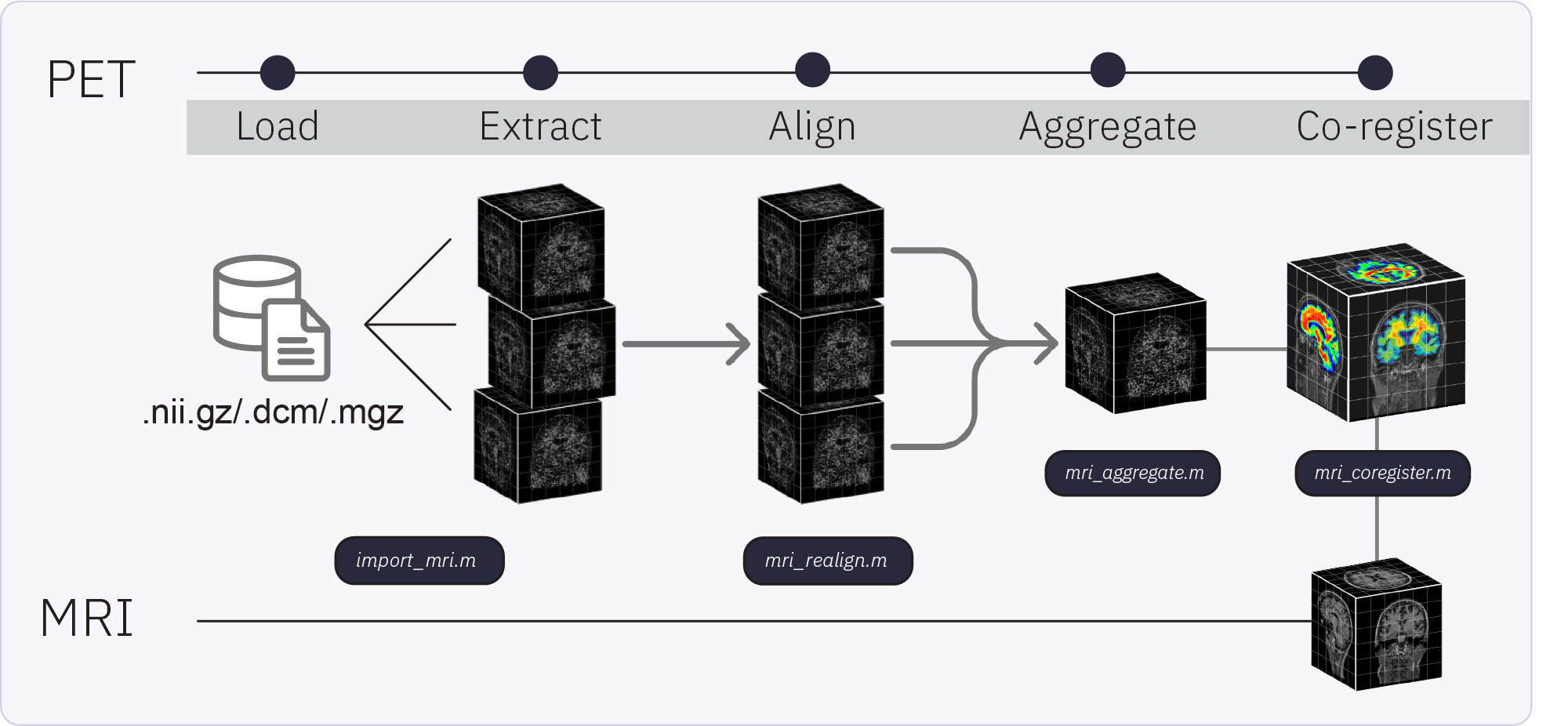
Dataset and requirements
Prerequisites:
Brainstorm Installation: Ensure you have a working copy of Brainstorm installed on your computer.
SPM12 (Statistical Parametric Mapping): required for realignment of dynamic (4D) PET volumes and co-registration of PET with MRI.
Import the anatomy
- Start Brainstorm
Select the menu File > Create new protocol. Name it "XXXXXX" and select the options:
"No, use individual anatomy",
"No, use one channel file per acquisition run".
Reference MRI
Go to the Anatomy view
Right-click on the XXXXXX top node > New subject > Subject01.
Keep the default options you defined for the protocol.Switch to the Anatomy view of the protocol.
Right-click on the subject node > Import MRI.
Set the file format: MRI: NIfTI-1 (*.nii;*.nii.gz)???.
Select: ???????.niiThe MRI viewer opens automatically. Click on "Click here to compute MNI normalization", option "maff8".
Click on Save to close the MRI viewer. New node named preMRI is created.
PET volume
The MRI volume above will be used as the anatomical reference for this subject. We will now import a PET scan done on the same subject. In this dataset, PET scan corresponds to XXXX.
Right-click on the subject node > Import PET.
Select: ????.niiChoose Yes for the transformation for MRI orientation.
- Choose the import options for PET
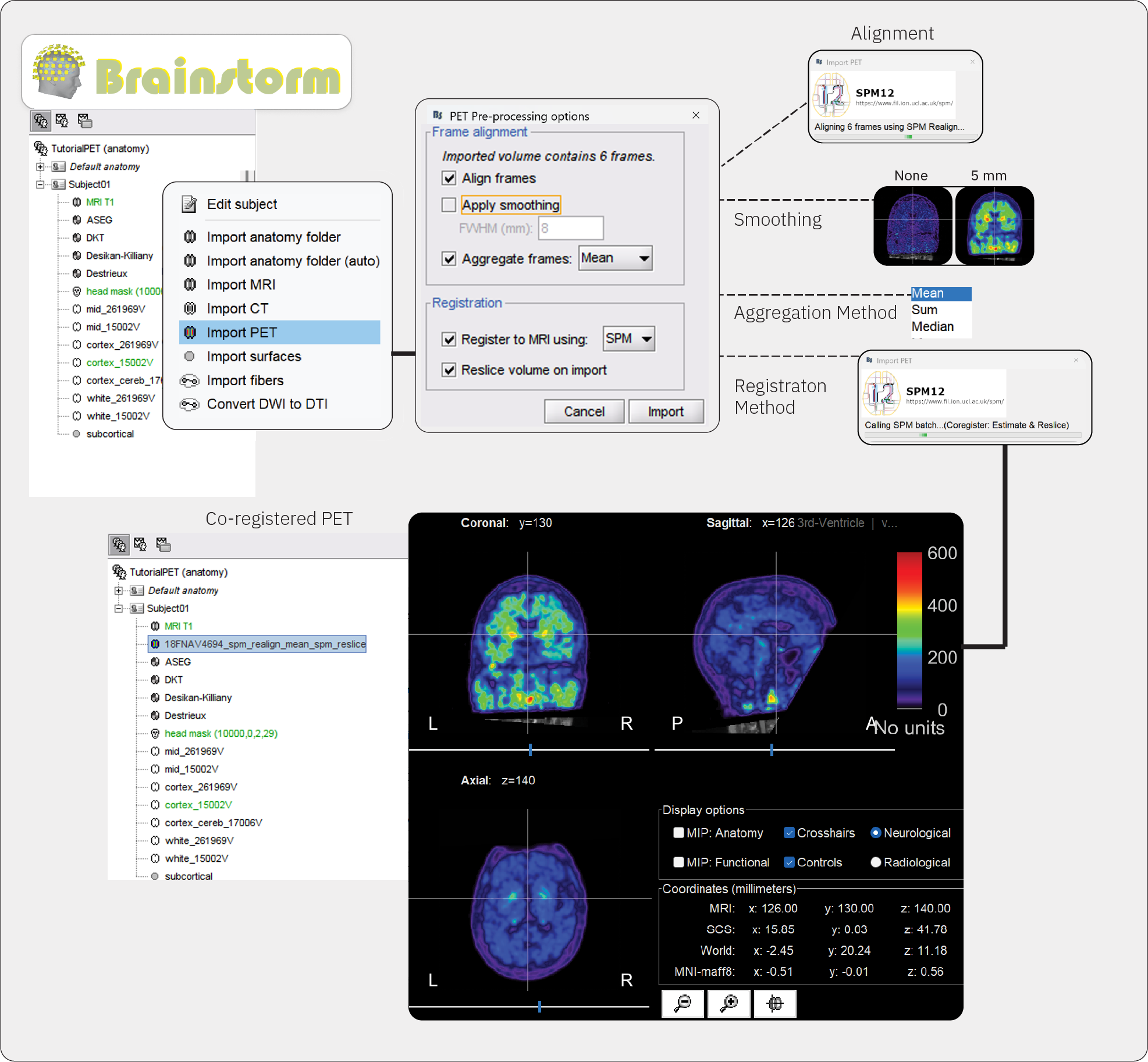
Align frames: Description on what happens, method, etc
Check this optionApply smoothing: Description
Check this option, and set FWHM to XX mmAggregate frames: Description
Select meanRegister to MRI using:: Description
Check this option, and set method to SPMReslice volume:: Description
Check this option
You can also decide to not perform any of these actions, and perform them once the PET volume is imported. IMAGE OF CONTEXT MENUS, for frame alignment and co-registration
![]() Edit conflict - other version:
Edit conflict - other version:
![]() Edit conflict - your version:
Edit conflict - your version:
![]() End of edit conflict
End of edit conflict
Surface-based analysis
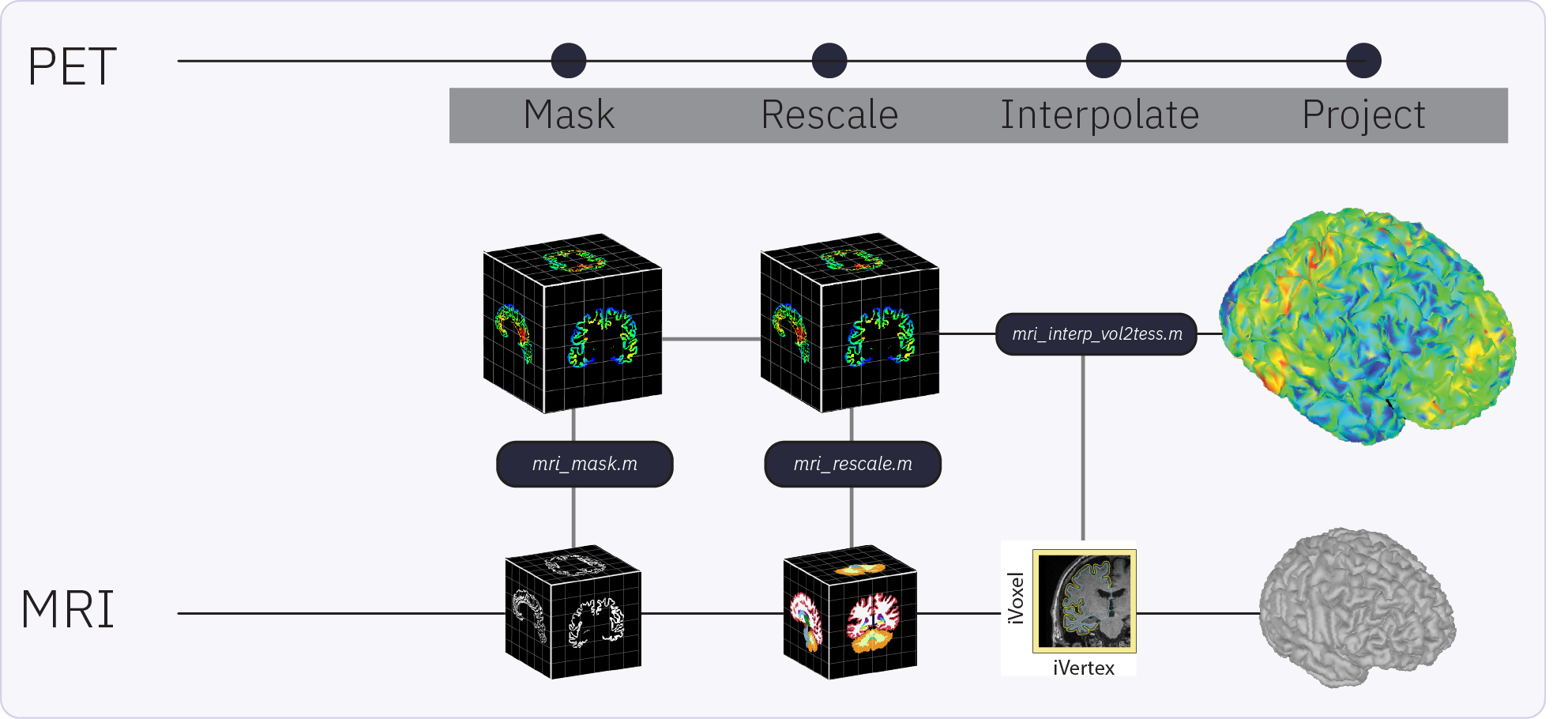
The surface-based submodule enables the projection of volumetric PET data onto the cortical surface, facilitating vertex-wise analysis in a common subject framework. Surface-based analysis of PET is based on the tess2mri interpolation matrix computed by Brainstorm. This matrix establishes weights for volumetric MRI voxels contributing to specific vertices on the cortical surface. Weights range between 0 (no contribution) and 1 (full contribution), enabling a smooth mapping of voxel coordinates to the cortical surface. Given that PET data are co-registered to the MRI, PET voxel intensity values are scaled by the interpolation weights, resulting in a weighted projection of PET values to the nearest vertex.
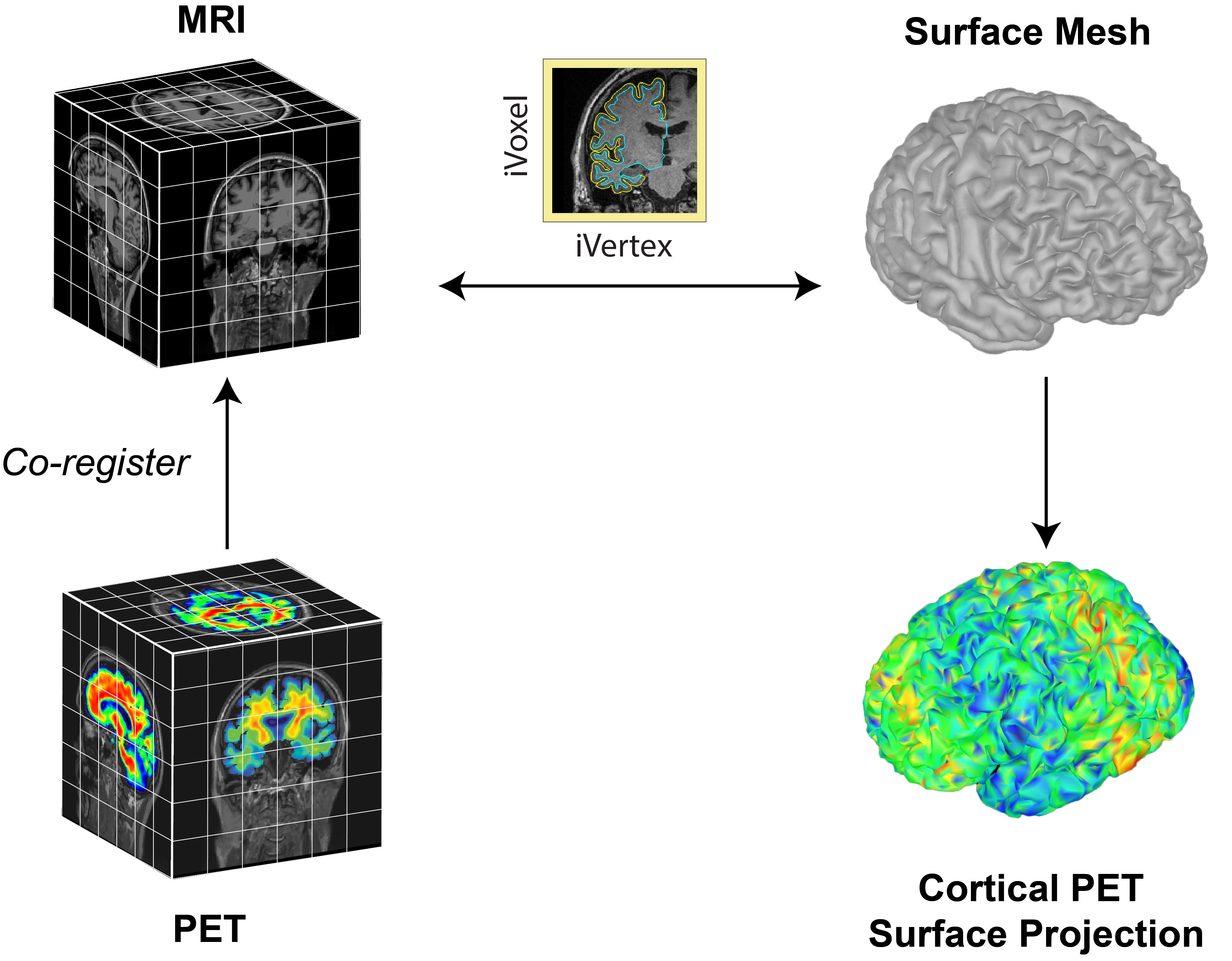
Related tutorials
Articles
Forum discussions
Overlaying PET on cortex: https://neuroimage.usc.edu/forums/t/30241
