MEG current phantom (CTF)
Authors: Francois Tadel, Elizabeth Bock
This tutorial explains how to use recordings from the CTF current phantom to test dipole fitting functions.
Contents
Phantom description
[TODO] Pictures
[TODO] Nature of the dipole that is generated
[TODO] References
Download and installation
Requirements: You have already followed all the introduction tutorials and you have a working copy of Brainstorm installed on your computer.
Go to the Download page of this website, and download the file: sample_phantom.zip
- Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder)
- Start Brainstorm (Matlab scripts or stand-alone version)
Select the menu File > Create new protocol. Name it "TutorialPhantom" and select the options:
"No, use individual anatomy",
"No, use one channel file per acquisition run (MEG)".
Generate anatomy
In the Matlab command window: type "generate_ctf_phantom".
This creates a new subject Phantom and generates the "anatomy" for this device: one volume and a few surfaces representing the geometry of the phantom.
You can display the MRI and surfaces as presented in the introduction tutorials.
Access the recordings
- Switch to the "functional data" view.
Right-click on the subject folder > Review raw file.
Select the file format: "MEG/EEG: CTF (*.ds)"
Select all the folders in: sample_phantom.
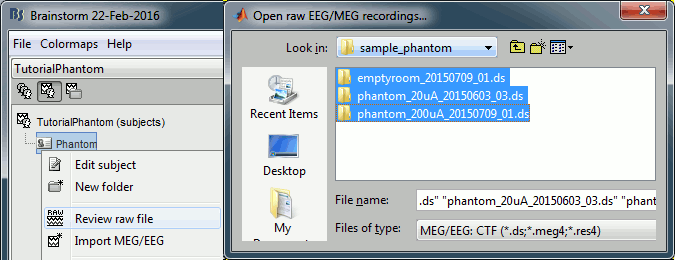
- Each folder corresponds to one dataset:
emptyroom_20150709_01.ds: Phantom inside the MEG helmet, but not plugged in
phantom_20uA_20150603_03.ds: Phantom active, 23Hz, 20uA, [0,-1.8,4.9]cm
phantom_200uA_20150709_01.ds: Phantom active, 7Hz, 200uA, [0,-1.8,4.9]cm
The recordings were acquired on different days, the position of the phantom in the MEG helmet is not the same for the two runs. Left = 20uA, Right = 200uA
For each of the three runs: right-click on "Link to raw file" > Switch epoched/continuous
Import recordings
Noise covariance
Source modeling
Dipole fitting with FieldTrip
Digitized head points
The head points collected with the Brainstorm digitizer are usually copied to the .ds folders and imported automatically when loading the recordings. We decided not to include them in this example because in the case of this current phantom, there is no ambiguity in the definition of the anatomical fiducials. As this refined registration with the .pos files is not part of the standard CTF workflow, it will make it easier to compare with other programs.
For additional testing purposes, the .pos file for the phantom is included in the sample_phantom.zaip package, but you have to add it manually to the recordings.
Right-click on one of the channel files (20uA or 200uA) >
Select the file format: "EEG: Polhemus"
Select file: sample_phantom/phantom_20160222_01.pos
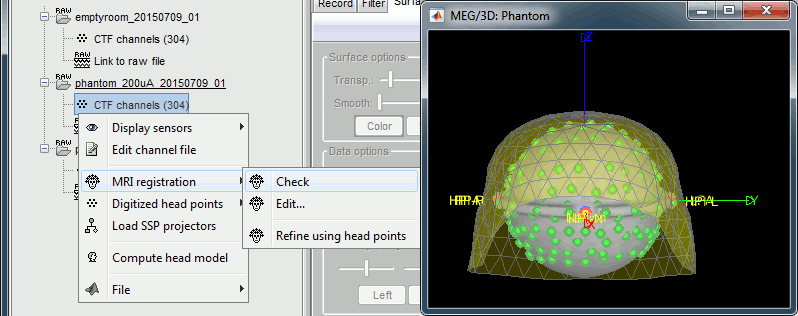
Right-click on the channel file > MRI registration > Check.
Scripting
Generate Matlab script
Available in the Brainstorm distribution: brainstorm3/toolbox/script/tutorial_phantom.m
