|
Size: 5959
Comment:
|
Size: 14080
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| http://neuroimage.usc.edu/brainstorm/Tutorials |
|
| Line 9: | Line 7: |
| Reviewers ''[current reviewing status] '' | Reviewers: ''[current reviewing status] '' |
| Line 11: | Line 9: |
| * '''Sylvain Baillet''': Montreal Neurological Institute ''[overview 1-14, no proofreading]'' * '''Richard Leahy''': University of Southern California ''[validated 1-19]'' * '''John Mosher''': Cleveland Clinic ''[not started]'' |
* '''Sylvain Baillet''': Montreal Neurological Institute ''[overview 1-14, edited 20-21]'' * '''Richard Leahy''': University of Southern California ''[validated 1-20]'' * '''John Mosher''': Cleveland Clinic ''[edited 22 only]'' |
| Line 15: | Line 13: |
Expected timeline (2016) * '''March''': Tutorials #1-#21 (interface and pre-processing) * '''April''': Tutorials #22-#24 (source estimation and time-frequency) * '''May-June''': Tutorials #25-#28 (statistics and workflows) * '''July-Sept''': Cross-validation of the pipeline and the results with MNE, FieldTrip and SPM * '''October''': Presentation during a satellite meeting at the Biomag 2016 conference * '''Nov-Dec''': Connectivity tutorial |
|
| Line 27: | Line 16: |
| == Tutorial 10: Power spectrum and frequency filters == | == Inverse models == === Tutorial 22: Source estimation === |
| Line 30: | Line 20: |
| * Richard, John, Sylvain: Finish the evaluation of the band-pass filters used in Brainstorm * Francois: Mark with an extended event the time segments that we cannot trust (edge effect) * Francois: Add warning if recordings are not long enough |
'''Note by John 2018/02/16''', after many discussions by phone and emails among Sylvain, Richard, Matti, Francois, and myself, the tentative decision is to release the modifications of the bst_inverse_linear_2016 as an updated code called "2018"; I'll leave it to Francois if this should be a new "bst_inverse_linear_2018" code, or simply change the Source Estimation panel to say "2018" using the older name. The two primary differences between 2016 and 2018 are that (1) 2018 now supports the mixed head model, such that "deep brain analysis" can run with 2018. (2) The other change to 2016 is an internal change in how noise is regularized. Matti Hamalainen (MNE-Python) and Rey (bst_wmne, our old Source Estimation) both chose the regularizer to be a fraction of the average of each sensor's variance (trace of the noise covariance divided by the number of sensors), which is the same as the average of the eigenvalues. So "0.1" as an input parameter in the "regularization" panel was calculated as 10% of the average variance. I used a different matrix norm, the maximum eigenvalue. Thus the same "0.1" would be 10% of the maximum eigenvalue, and given the large dynamic range of MEG sensors, this is a substantial difference in regularizers between the codes. Thus user's using the same fraction in both methods saw disparate results. We made the decision to change the "reg" option to be nearly the same as Rey's code, to lessen confusion among users switching between codes and platforms. We need to also strengthen the general discussion on the importance of noise regularization. The new 2018 is "nearly" the same as Rey/Matti, because there is still an open debate on what to do with the cross-correlation terms between modalities. Matti is double checking his codes, but Rey's interpretation/implementation in 2011 was that the cross terms between, say, EEG and MEG, were zero'd out. In Source 2018, I continue this philosophy by doing the same between GRADS and MAGS, which apparently Rey/Matti do keep the cross terms. After conceptual discussions with Richard, we decided to be conservative and zero out the cross terms between all modalities. I have coded 2018 to make this an easily adjustable flag that can be later tested and distributed, if we desire. Two other fixes in 2018 were made related to the above. (1) 2016 was not regularizing correctly across multiple modalities, by trying to find a single regularizer to the overall matrix. Thus multiple modalities did not work well, since one modality tended to dominate the eigenspectrum and the other modalities were ignored. Each modalitiey is now separately regularized by all of the regularization methods (GRADS and MAGS as well), then recombined back into an overall noise covariance matrix for joint estimation. The open question, discussed above, is whether or not to put the cross terms back between modalities back into this matrix, and we have turned that OFF for now (Feb 2018). (2) Once the regularizer value "lambda" was selected, I had a bug in that I formed sqrt(eigenvalue)+sqrt(lambda), rather than the correct sqrt(eigenvalue + lambda). So that users understand that they are running a new version of source estimation that may yield different results from earlier, we decided that it would be better to call this "Source Estimation 2018", rather than e.g. 2016 (fixed). So the below comments that say "fixed in 2018" reference the above discussion. * '''John''': Inverse code: Mixed head models are still not supported. '''Fixed in 2018 ''' * '''John''': Explain the new (incorrect) results obtained with the epilepsy tutorial (see below) '''Fixed in 2018,''' the issue was that the estimate was over-regularized in the new version, when using the same regularization parameter. * '''John''': Explain the donut shapes of the min norm maps we always get with this new function. '''Probably Fixed in 2018''', since I don't see these donuts, and my min norms look like the old min norms, once we equate regularization parameters. * '''John''': Drop the option "RMS source amplitude"? '''As time permits, okay, can be dropped. '''I discussed this with Richard and Matti. We currently regularize based on an SNR ratio, that's frankly pulled out of the air, e.g. "3". There is another way, based on the physiologic concept of the expected signal strength, such as the "Okada constant" (1 nA-m/mm max possible). But Matti agrees this remains untested. Richard wonders if it's okay to just leave it dormant in the code. Francois would like to clean up the interfaces and codes. So basically, Francois, you're free to remove it for now, we'll return to this alternative in later years, maybe from a different viewpoint. * * The option is not even accessible in the interface: you successively asked me to disable it for the min norm, and then made me hide the entire section "Regularization parameter" for the dipole modelling and the beamformer. * Can I just remove it from the interface? * Francois: Update code, tutorials and screen captures accordingly * * '''John, Richard, Sylvain, Matti, Alex''': Make the "median eigenvalue" option the default? * . '''Addressed somewhat by 2018 code''', but this is still a professional opinion. MEG data in thinly shielded rooms have an enormous dynamic range. Simply taking 10% of the trace average or even 1% of the matrix norm is inadequate to capture properly the truly smaller yet valid eigenvalues. We want regularization to fix deficiently small eigenvalue deep in the tail, but the "reg" option is too crude. In conversation with Matti, for example, he doesn't even use this option with his data, because his mammoth shielded room is that good. So I see no problem in writing that I (Mosher) recommend median eigenvalue, while the code and the user want to try something else. * John suggests to use the "median eigenvalue" option by default instead of the option "Regularize noise covariance", which as been used for many years. * In [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Advanced_options:_Minimum_norm|this section]] of the tutorials, John wrote: "'''Recommended option''': This author (Mosher) votes for the '''median eigenvalue '''as being generally effective. The other options are useful for comparing with other software packages that generally employ similar regularization methods." * However this modifies a lot the results: the localization results and the MN amplitudes can be very different. If this is a clear improvement, it's good to promote it. But it cannot be done randomly like this, this has to be discussed (especially with Matti and Alex) and tested. * John: Please arrange a meeting so you can discuss this question. * '''John, Richard, Sylvain''': Why are dSPM values 2x lower than Z-score ? * '''Feb 2018:''' This remains a bit of a mystery. In separate conversations, Mosher, Hamalainen, and Leahy each think in principle there should be no difference; however, they also acknowledge that the difference may lie in how the cross covariance vs auto-covariance is utilized in either or both. As discussed above, in wide dynamic range such as MEG, there is an enormous difference between using the diagonal values of the noise covariance matrix vs. the full cross-covariance, so does the problem lay here, where one or the other method is not fully exploiting the cross terms? We need a good representative test case to explore. * The tutorial says "Z-normalized current density maps are also easy to interpret. They represent explicitly a "deviation from experimental baseline" as defined by the user. In contrast, dSPM indicates the deviation from the data that was used to define the noise covariance used in computing the min norm map. " * Therefore should we expect the dSPM values to deviate more from the noise recordings, than the Z-score from the pre-stim baseline? Instead of this we observe much lower values. Is there a scaling issue here? <<BR>><<BR>> {{attachment:diff_zscore_dspm.gif||height="138",width="385"}} * '''John''': Mixing GRAD and MAG: * '''John''': You do not recommend processing GRAD and MAG at the same time? This is currently the default behavior in the interface... '''Fixed in 2018 '''you can use either or both, since we have fixed the regularization issues in cross-modalities. * '''John''': Please discuss this with Matti and Alex * '''Francois''': Change the default + add note in Elekta tutorial if change is validated * '''Francois''': Call FieldTrip headmodels and beamformers |
| Line 36: | Line 63: |
| * Richard, John, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsFilter#What_filters_to_apply.3F_.5BTODO.5D|What filters to apply?]] * Richard, John, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsFilter#Filters_specifications_.5BTODO.5D|Filters specifications]] |
* '''John''': Fix all the missing links '''Feb 2018, working. ''' These were actually Richard's links, so I'm reverse engineering what he was thinking about crosslinking to. * '''John''': Data covariance: * . '''Feb 2018: '''I (John) recommend you (Francois) just leave the defaults alone for now. We are working on a new set of papers addressing the beamformers. As Richard has also learned, there is a lot of confusion on how much and what you actually feed a so-called beamformer. |
| Line 39: | Line 67: |
| == Tutorial 12: Artifact detection == [ONLINE DOC] |
* Recommendations moved to the [[http://neuroimage.usc.edu/brainstorm/Tutorials/NoiseCovariance#Data_covariance|Noise and data covariance tutorial]]. * You said: "Our recommendation for evoked responses is to use a window that spans prestim through the end of the response of interest, with a minimum of 500ms total duration. " * Should I modify the interface (and screen capture of the example) to always include the pre-stim baseline (eg. from -100ms to +500ms, instead of from 0ms to +500ms) ? * '''Francois''': Update the screen capture + code for default selection of the time window |
| Line 42: | Line 72: |
| * Beth: Adding examples of the different detection processes. Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsDetect#Other_detection_processes|Other detection processes]] | === Tutorial: Dipole scanning === * '''John''': Unfinished sentence in [[http://neuroimage.usc.edu/brainstorm/Tutorials/TutDipScan#Dipole_information|this section]]. |
| Line 44: | Line 75: |
| == Tutorial 13: Artifact cleaning with SSP == | === Tutorial EEG/Epilepsy === * '''John''': Why sLORETA? '''Feb 2018: '''sLORETA is a favorite among some of the epilepsy community, particularly Europeans. When processing spikes as single sources, we (Hamalainen and I) do not expect much practical difference between the dSPM and sLORETA. No reason to poke that bear. * '''John''': Please address the location issues with the new code: <<BR>>dSPM is now localizing the spike in a much deeper spot (top=old version, bottom=new version) <<BR>><<BR>> {{attachment:epilepsy_dspm.gif||height="233",width="272"}} * '''Marcel '''wrote: "Looking at the findings from the intracranial EEG in Matthias Dümpelmann's article (figure 1 panel a) and b)) it is '''very likely the new sources are wrong'''. Additionally, the older sources are much more in agreement with Matthias' sLORETA results and with cMEM findings. Sohrabopour et al. reported as well that their IRES method found results in agreement with sources shown in the tutorial." * Imported data for testing can be downloaded here:<<BR>> https://www.dropbox.com/s/42d9indpjr8ac1y/TutorialEpilepsy.zip?dl=0 * '''Fixed in 2018:''' The clue lies in the z-scores of the two DSPM images. The bottom one (new code) is much suppressed (peak of 12), because it was over-regularized, compared the upper one (peak of 25). In the 2018 code, the same "0.1" parameter is now used equivalently to be the fraction of the average eigenvalue, i.e. 10% of the average sensor variance. == Filters == === Tutorial 10: Power spectrum and frequency filters === |
| Line 47: | Line 86: |
| * Francois: Check the length needed to filter the recordings (after finishing #10) <<BR>>Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsSsp#SSP_Algorithm_.5BTODO.5D|SSP Algorithm]] == Tutorial 15: Import epochs == [ONLINE DOC] * Richard, Sylvain, John, Francois: Define recommendations for epoch lengths (after finishing #10) <<BR>>Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epoching#Epoch_length_.5BTODO.5D|Epoch length]] == Tutorial 20: Head modelling == [ONLINE DOC] * John, Richard, Sylvain: In-depth review of this sensitive tutorial * John, Richard, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/AllIntroduction#Tutorials.2FHeadModel.References_.5BTODO.5D|References]] == Tutorial 22: Source estimation == [CODE] * John: Dipole scanning: Wrong orientations * John: Inverse code: sLORETA * John: Inverse code: NAI * John: Inverse code: Mixed head models * John, Richard, Sylvain: How to deal with '''unconstrained sources''' ? * For the Z-score normalization? * For the connectivity analysis? http://neuroimage.usc.edu/forums/showthread.php?2401 * For the statistics? * Projection on a dominant orientation? * Francois: Remove the warning messages in the interface * Francois: Call FieldTrip headmodels and beamformers |
* '''Hossein, Francois''': Update the code for the band-stop and notch filters in the same way * Look at code in each case and write additional code to compute equivalent impulse response and give transient duration (99%) energy info as well as frequency and impulse response plots to user as we do with the bandpass/lowpass filters. |
| Line 77: | Line 91: |
| * John, Richard, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Source_estimation_options_.5BTODO.5D|Source estimation options]] * John, Richard, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Advanced_options_.5BTODO.5D|Advanced options]] * John, Richard, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Equations_.5BTODO.5D|Equations]] * John, Richard, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#References_.5BTODO.5D|References]] * John, Richard, Sylvain: Why are dSPM values 2x lower than Z-score ? * John, Richard, Sylvain: In-depth review of this sensitive tutorial |
* '''Hossein, Francois''': Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsFilter#Filters_specifications_.5BTODO.5D|Filters specifications]] for band-stop and notch filters |
| Line 84: | Line 93: |
| == Tutorial 24: Time-frequency == [CODE] * Enable option "Hide edge effects" for Hilbert (after finishing #10) [ONLINE DOC] * Hilbert: Link back to the filters tutorial (after finishing #10) == Tutorial 25: Difference == [ONLINE DOC] * Validate order of the processes: Difference sources => Zscore => Low-pass filter<<BR>>Section: [[http://neuroimage.usc.edu/brainstorm/Tutorials/Difference#Difference_deviant-standard|Difference deviant-standard]] == Tutorial 26: Statistics == * Not evaluated yet |
* '''Francois''': Update section "Apply a notch filter" to add the transients in screen capture and text (after the documentation of the notch filter) |
| Line 102: | Line 96: |
| * Not evaluated yet * Update: http://neuroimage.usc.edu/brainstorm/Tutorials/WorkflowGuide |
* Add Chi2(log) ? |
| Line 105: | Line 98: |
| == Tutorial 28: Scripting == * Not evaluated yet == Final steps == * Francois: Check that the script tutorial_introduction.m produces the same output as tutorials * Francois: Check all the links in all the pages * Francois: Check the number of pages for each tutorial * Francois: Reference on ResearchGate, Academia and Google Scholar <<BR>>http://neuroimage.usc.edu/brainstorm/Tutorials/AllIntroduction * Francois: Remove useless images from all tutorials <<BR>><<BR>><<BR>><<BR>><<BR>>[Additional important stuff] == Phantom tutorial (for validation) == * John: Prepare Elekta recordings to distribute on the [[Tutorials/PhantomElekta|tutorial page]] * Francois: Prepare the Elekta phantom anatomy * Francois: Prepare the analysis script * Francois: Edit the tutorial page == Other analysis scenarios == * Francois: Update all the tutorials (100+ pages) * Francois: Add number of pages in the tutorials |
== Group study tutorial == * Statistics must be validated and results must be explained properly. * http://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup#Group_analysis:_Sources |
| Line 129: | Line 104: |
| * Richard, Sylvain: Define example dataset and precise results to obtain from them * Richard: How to assess significance from connectivity matrices * Francois: Preparation of a tutorial |
* '''Richard, Sylvain''': Define example dataset and precise results to obtain from them * '''Richard, Sylvain''': How to deal with unconstrained sources?<<BR>> http://neuroimage.usc.edu/forums/showthread.php?2401 * '''Richard''': How to assess significance from connectivity matrices? * '''Richard, Hossein, Francois''': Preparation of a tutorial * All the functions using bandpass filters must be updated to use the new filters == Final steps == * Francois: Remove all the wiki pages that are not used * Francois: Check all the links in all the pages * Francois: Check that all the TODO blocks have been properly handled * Francois: Remove useless images from all tutorials * Francois: Update page count on the main tutorials page * Francois: Reference on ResearchGate, Academia and Google Scholar <<BR>>http://neuroimage.usc.edu/brainstorm/Tutorials/AllIntroduction |
Introduction tutorials: Editing process
Redactors:
Francois Tadel: Montreal Neurological Institute
Elizabeth Bock: Montreal Neurological Institute
Reviewers: [current reviewing status]
Sylvain Baillet: Montreal Neurological Institute [overview 1-14, edited 20-21]
Richard Leahy: University of Southern California [validated 1-20]
John Mosher: Cleveland Clinic [edited 22 only]
Dimitrios Pantazis: Massachusetts Institute of Technology [validated 1-15]
Inverse models
Tutorial 22: Source estimation
[CODE]
Note by John 2018/02/16, after many discussions by phone and emails among Sylvain, Richard, Matti, Francois, and myself, the tentative decision is to release the modifications of the bst_inverse_linear_2016 as an updated code called "2018"; I'll leave it to Francois if this should be a new "bst_inverse_linear_2018" code, or simply change the Source Estimation panel to say "2018" using the older name.
The two primary differences between 2016 and 2018 are that (1) 2018 now supports the mixed head model, such that "deep brain analysis" can run with 2018.
(2) The other change to 2016 is an internal change in how noise is regularized. Matti Hamalainen (MNE-Python) and Rey (bst_wmne, our old Source Estimation) both chose the regularizer to be a fraction of the average of each sensor's variance (trace of the noise covariance divided by the number of sensors), which is the same as the average of the eigenvalues. So "0.1" as an input parameter in the "regularization" panel was calculated as 10% of the average variance. I used a different matrix norm, the maximum eigenvalue. Thus the same "0.1" would be 10% of the maximum eigenvalue, and given the large dynamic range of MEG sensors, this is a substantial difference in regularizers between the codes. Thus user's using the same fraction in both methods saw disparate results. We made the decision to change the "reg" option to be nearly the same as Rey's code, to lessen confusion among users switching between codes and platforms. We need to also strengthen the general discussion on the importance of noise regularization.
The new 2018 is "nearly" the same as Rey/Matti, because there is still an open debate on what to do with the cross-correlation terms between modalities. Matti is double checking his codes, but Rey's interpretation/implementation in 2011 was that the cross terms between, say, EEG and MEG, were zero'd out. In Source 2018, I continue this philosophy by doing the same between GRADS and MAGS, which apparently Rey/Matti do keep the cross terms. After conceptual discussions with Richard, we decided to be conservative and zero out the cross terms between all modalities. I have coded 2018 to make this an easily adjustable flag that can be later tested and distributed, if we desire.
Two other fixes in 2018 were made related to the above. (1) 2016 was not regularizing correctly across multiple modalities, by trying to find a single regularizer to the overall matrix. Thus multiple modalities did not work well, since one modality tended to dominate the eigenspectrum and the other modalities were ignored. Each modalitiey is now separately regularized by all of the regularization methods (GRADS and MAGS as well), then recombined back into an overall noise covariance matrix for joint estimation. The open question, discussed above, is whether or not to put the cross terms back between modalities back into this matrix, and we have turned that OFF for now (Feb 2018).
(2) Once the regularizer value "lambda" was selected, I had a bug in that I formed sqrt(eigenvalue)+sqrt(lambda), rather than the correct sqrt(eigenvalue + lambda).
So that users understand that they are running a new version of source estimation that may yield different results from earlier, we decided that it would be better to call this "Source Estimation 2018", rather than e.g. 2016 (fixed).
So the below comments that say "fixed in 2018" reference the above discussion.
John: Inverse code: Mixed head models are still not supported. Fixed in 2018
John: Explain the new (incorrect) results obtained with the epilepsy tutorial (see below) Fixed in 2018, the issue was that the estimate was over-regularized in the new version, when using the same regularization parameter.
John: Explain the donut shapes of the min norm maps we always get with this new function. Probably Fixed in 2018, since I don't see these donuts, and my min norms look like the old min norms, once we equate regularization parameters.
John: Drop the option "RMS source amplitude"? As time permits, okay, can be dropped. I discussed this with Richard and Matti. We currently regularize based on an SNR ratio, that's frankly pulled out of the air, e.g. "3". There is another way, based on the physiologic concept of the expected signal strength, such as the "Okada constant" (1 nA-m/mm max possible). But Matti agrees this remains untested. Richard wonders if it's okay to just leave it dormant in the code. Francois would like to clean up the interfaces and codes. So basically, Francois, you're free to remove it for now, we'll return to this alternative in later years, maybe from a different viewpoint.
- * The option is not even accessible in the interface: you successively asked me to disable it for the min norm, and then made me hide the entire section "Regularization parameter" for the dipole modelling and the beamformer.
- Can I just remove it from the interface?
- Francois: Update code, tutorials and screen captures accordingly
John, Richard, Sylvain, Matti, Alex: Make the "median eigenvalue" option the default?
. Addressed somewhat by 2018 code, but this is still a professional opinion. MEG data in thinly shielded rooms have an enormous dynamic range. Simply taking 10% of the trace average or even 1% of the matrix norm is inadequate to capture properly the truly smaller yet valid eigenvalues. We want regularization to fix deficiently small eigenvalue deep in the tail, but the "reg" option is too crude. In conversation with Matti, for example, he doesn't even use this option with his data, because his mammoth shielded room is that good. So I see no problem in writing that I (Mosher) recommend median eigenvalue, while the code and the user want to try something else.
- John suggests to use the "median eigenvalue" option by default instead of the option "Regularize noise covariance", which as been used for many years.
In this section of the tutorials, John wrote: "Recommended option: This author (Mosher) votes for the median eigenvalue as being generally effective. The other options are useful for comparing with other software packages that generally employ similar regularization methods."
- However this modifies a lot the results: the localization results and the MN amplitudes can be very different. If this is a clear improvement, it's good to promote it. But it cannot be done randomly like this, this has to be discussed (especially with Matti and Alex) and tested.
- John: Please arrange a meeting so you can discuss this question.
John, Richard, Sylvain: Why are dSPM values 2x lower than Z-score ?
Feb 2018: This remains a bit of a mystery. In separate conversations, Mosher, Hamalainen, and Leahy each think in principle there should be no difference; however, they also acknowledge that the difference may lie in how the cross covariance vs auto-covariance is utilized in either or both. As discussed above, in wide dynamic range such as MEG, there is an enormous difference between using the diagonal values of the noise covariance matrix vs. the full cross-covariance, so does the problem lay here, where one or the other method is not fully exploiting the cross terms? We need a good representative test case to explore.
- The tutorial says "Z-normalized current density maps are also easy to interpret. They represent explicitly a "deviation from experimental baseline" as defined by the user. In contrast, dSPM indicates the deviation from the data that was used to define the noise covariance used in computing the min norm map. "
Therefore should we expect the dSPM values to deviate more from the noise recordings, than the Z-score from the pre-stim baseline? Instead of this we observe much lower values. Is there a scaling issue here?
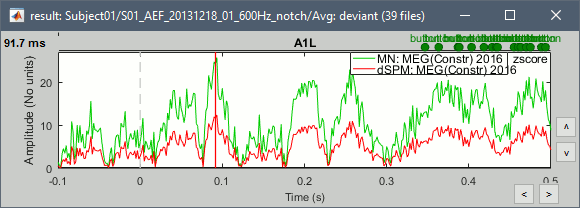
John: Mixing GRAD and MAG:
John: You do not recommend processing GRAD and MAG at the same time? This is currently the default behavior in the interface... Fixed in 2018 you can use either or both, since we have fixed the regularization issues in cross-modalities.
John: Please discuss this with Matti and Alex
Francois: Change the default + add note in Elekta tutorial if change is validated
Francois: Call FieldTrip headmodels and beamformers
[ONLINE DOC]
John: Fix all the missing links Feb 2018, working. These were actually Richard's links, so I'm reverse engineering what he was thinking about crosslinking to.
John: Data covariance:
. Feb 2018: I (John) recommend you (Francois) just leave the defaults alone for now. We are working on a new set of papers addressing the beamformers. As Richard has also learned, there is a lot of confusion on how much and what you actually feed a so-called beamformer.
Recommendations moved to the Noise and data covariance tutorial.
- You said: "Our recommendation for evoked responses is to use a window that spans prestim through the end of the response of interest, with a minimum of 500ms total duration. "
- Should I modify the interface (and screen capture of the example) to always include the pre-stim baseline (eg. from -100ms to +500ms, instead of from 0ms to +500ms) ?
Francois: Update the screen capture + code for default selection of the time window
Tutorial: Dipole scanning
John: Unfinished sentence in this section.
Tutorial EEG/Epilepsy
John: Why sLORETA? Feb 2018: sLORETA is a favorite among some of the epilepsy community, particularly Europeans. When processing spikes as single sources, we (Hamalainen and I) do not expect much practical difference between the dSPM and sLORETA. No reason to poke that bear.
John: Please address the location issues with the new code:
dSPM is now localizing the spike in a much deeper spot (top=old version, bottom=new version)
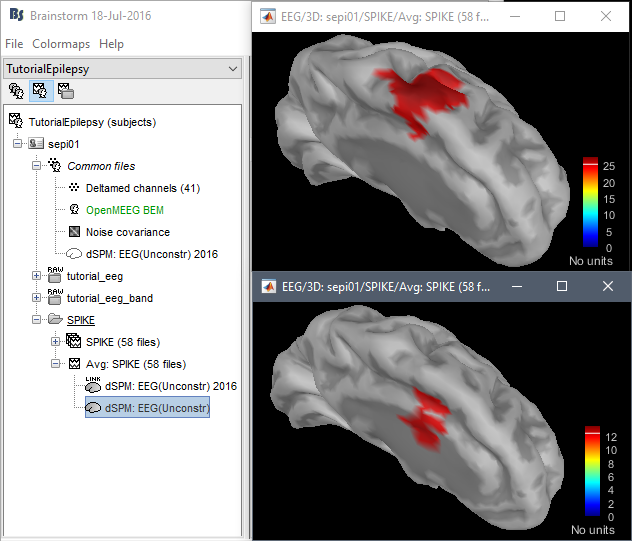
Marcel wrote: "Looking at the findings from the intracranial EEG in Matthias Dümpelmann's article (figure 1 panel a) and b)) it is very likely the new sources are wrong. Additionally, the older sources are much more in agreement with Matthias' sLORETA results and with cMEM findings. Sohrabopour et al. reported as well that their IRES method found results in agreement with sources shown in the tutorial."
Imported data for testing can be downloaded here:
https://www.dropbox.com/s/42d9indpjr8ac1y/TutorialEpilepsy.zip?dl=0Fixed in 2018: The clue lies in the z-scores of the two DSPM images. The bottom one (new code) is much suppressed (peak of 12), because it was over-regularized, compared the upper one (peak of 25). In the 2018 code, the same "0.1" parameter is now used equivalently to be the fraction of the average eigenvalue, i.e. 10% of the average sensor variance.
Filters
Tutorial 10: Power spectrum and frequency filters
[CODE]
Hossein, Francois: Update the code for the band-stop and notch filters in the same way
- Look at code in each case and write additional code to compute equivalent impulse response and give transient duration (99%) energy info as well as frequency and impulse response plots to user as we do with the bandpass/lowpass filters.
[ONLINE DOC]
Hossein, Francois: Section Filters specifications for band-stop and notch filters
Francois: Update section "Apply a notch filter" to add the transients in screen capture and text (after the documentation of the notch filter)
Tutorial 27: Workflows
- Add Chi2(log) ?
Group study tutorial
- Statistics must be validated and results must be explained properly.
http://neuroimage.usc.edu/brainstorm/Tutorials/VisualGroup#Group_analysis:_Sources
Connectivity
- Not documented at all
Richard, Sylvain: Define example dataset and precise results to obtain from them
Richard, Sylvain: How to deal with unconstrained sources?
http://neuroimage.usc.edu/forums/showthread.php?2401Richard: How to assess significance from connectivity matrices?
Richard, Hossein, Francois: Preparation of a tutorial
- All the functions using bandpass filters must be updated to use the new filters
Final steps
- Francois: Remove all the wiki pages that are not used
- Francois: Check all the links in all the pages
- Francois: Check that all the TODO blocks have been properly handled
- Francois: Remove useless images from all tutorials
- Francois: Update page count on the main tutorials page
Francois: Reference on ResearchGate, Academia and Google Scholar
http://neuroimage.usc.edu/brainstorm/Tutorials/AllIntroduction
