|
Size: 7516
Comment:
|
Size: 7474
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 9: | Line 9: |
| Reviewers ''[current reviewing status] '' | Reviewers: ''[current reviewing status] '' |
| Line 11: | Line 11: |
| * '''Sylvain Baillet''': Montreal Neurological Institute ''[overview 1-14, no proofreading]'' * '''Richard Leahy''': University of Southern California ''[validated 1-19]'' * '''John Mosher''': Cleveland Clinic ''[not started]'' |
* '''Sylvain Baillet''': Montreal Neurological Institute ''[overview 1-14, edited 20-21]'' * '''Richard Leahy''': University of Southern California ''[validated 1-20]'' * '''John Mosher''': Cleveland Clinic ''[edited 22 only]'' |
| Line 15: | Line 15: |
Expected timeline (2016) * '''March''': Tutorials #1-#21 (interface and pre-processing) * '''April''': Tutorials #22-#24 (source estimation and time-frequency) * '''May-June''': Tutorials #25-#28 (statistics and workflows) * '''July-Sept''': Cross-validation of the pipeline and the results with MNE, FieldTrip and SPM * '''October''': Presentation during a satellite meeting at the Biomag 2016 conference * '''Nov-Dec''': Connectivity tutorial |
|
| Line 27: | Line 18: |
| == Tutorial 10: Power spectrum and frequency filters == | == Inverse models == === Tutorial 22: Source estimation === |
| Line 30: | Line 22: |
| * Richard, John, Sylvain: Finish the evaluation of the band-pass filters used in Brainstorm * Francois: Mark with an extended event the time segments that we cannot trust (edge effect) * Francois: Add warning if recordings are not long enough [ONLINE DOC] * Richard, John, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsFilter#What_filters_to_apply.3F_.5BTODO.5D|What filters to apply?]] * Richard, John, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsFilter#Filters_specifications_.5BTODO.5D|Filters specifications]] == Tutorial 12: Artifact detection == [ONLINE DOC] * Beth: Adding examples of the different detection processes. Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsDetect#Other_detection_processes|Other detection processes]] == Tutorial 13: Artifact cleaning with SSP == [CODE] * Francois: Check the length needed to filter the recordings (after finishing #10) <<BR>>Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsSsp#SSP_Algorithm_.5BTODO.5D|SSP Algorithm]] == Tutorial 15: Import epochs == [ONLINE DOC] * Richard, Sylvain, John, Francois: Define recommendations for epoch lengths (after finishing #10) <<BR>>Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epoching#Epoch_length_.5BTODO.5D|Epoch length]] == Tutorial 20: Head modelling == [ONLINE DOC] * John, Richard, Sylvain: In-depth review of this sensitive tutorial * John, Richard, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/AllIntroduction#Tutorials.2FHeadModel.References_.5BTODO.5D|References]] == Tutorial 22: Source estimation == [CODE] * John: Inverse code: sLORETA * John: Inverse code: NAI * John: Inverse code: Mixed head models * John, Richard, Sylvain: How to deal with '''unconstrained sources''' ? * For the Z-score normalization? * For the connectivity analysis? http://neuroimage.usc.edu/forums/showthread.php?2401 * For the statistics? * Projection on a dominant orientation? * Francois: Remove the warning messages in the interface |
* '''John''': Inverse code: Mixed head models are still not supported. * '''John''': Explain the new (incorrect) results obtained with the epilepsy tutorial (see below) * '''John''': Drop the option "RMS source amplitude"? * The documentation is not informative and not encouraging at all: "RMS source amplitude: An alternative definition of SNR, but still under test and may be dropped." * The option is not even accessible in the interface: you successively asked me to disable it for the min norm, and then made me hide the entire section "Regularization parameter" for the dipole modelling and the beamformer. * Can I just remove it from the interface? * Francois: Update code, tutorials and screen captures accordingly * '''John, Richard, Sylvain, Matti, Alex''': Make the "median eigenvalue" option the default? * John suggests to use the "median eigenvalue" option by default instead of the option "Regularize noise covariance", which as been used for many years. * In [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Advanced_options:_Minimum_norm|this section]] of the tutorials, John wrote: "'''Recommended option''': This author (Mosher) votes for the '''median eigenvalue '''as being generally effective. The other options are useful for comparing with other software packages that generally employ similar regularization methods." * However this modifies a lot the results: the localization results and the MN amplitudes can be very different. If this is a clear improvement, it's good to promote it. But it cannot be done randomly like this, this has to be discussed (especially with Matti and Alex) and tested. * John: Please arrange a meeting so you can discuss this question. * '''John, Richard, Sylvain''': Why are dSPM values 2x lower than Z-score ? * The tutorial says "Z-normalized current density maps are also easy to interpret. They represent explicitly a "deviation from experimental baseline" as defined by the user. In contrast, dSPM indicates the deviation from the data that was used to define the noise covariance used in computing the min norm map. " * Therefore should we expect the dSPM values to deviate more from the noise recordings, than the Z-score from the pre-stim baseline? Instead of this we observe much lower values. Is there a scaling issue here? <<BR>><<BR>> {{attachment:diff_zscore_dspm.gif||width="385",height="138"}} * Mixing GRAD and MAG: * '''John''': You do not recommend processing GRAD and MAG at the same time? This is currently the default behavior in the interface... * '''John''': Please discuss this with Matti and Alex * Francois: Change the default + add note in Elekta tutorial if change is validated * '''John''': Send a message to Margot Taylor: She's been asking for your beamformers for about 2yrs. |
| Line 76: | Line 46: |
| * John, Richard, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Source_estimation_options_.5BTODO.5D|Source estimation options]] * John, Richard, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Advanced_options_.5BTODO.5D|Advanced options]] * John, Richard, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Equations_.5BTODO.5D|Equations]] * John, Richard, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#References_.5BTODO.5D|References]] * John, Richard, Sylvain: Why are dSPM values 2x lower than Z-score ? * John, Richard, Sylvain: In-depth review of this sensitive tutorial |
* '''John''': Fix all the missing links * '''John, Richard, Sylvain''': Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#References_.5BTODO.5D|References]] * '''John, Richard, Sylvain''': What do with [[http://neuroimage.usc.edu/brainstorm/Tutorials/Beamformers|Hui-Ling Beamformers]]? === Tutorial: Dipole scanning === * '''John''': Add the description of all the measures in [[http://neuroimage.usc.edu/brainstorm/Tutorials/TutDipScan#Significance_mesures_.5BTODO.5D|this section]]. === Tutorial EEG/Epilepsy === * '''John''': Why sLORETA? * '''John''': Please address the location issues with the new code: <<BR>>dSPM is now localizing the spike in a much deeper spot (top=old version, bottom=new version) <<BR>><<BR>> {{attachment:epilepsy_dspm.gif||width="272",height="233"}} * '''Marcel Heers''' wrote: "Looking at the findings from the intracranial EEG in Matthias Dümpelmann's article (figure 1 panel a) and b)) it is '''very likely the new sources are wrong'''. Additionally, the older sources are much more in agreement with Matthias' sLORETA results and with cMEM findings. Sohrabopour et al. reported as well that their IRES method found results in agreement with sources shown in the tutorial." * Imported data for testing can be downloaded here:<<BR>> https://www.dropbox.com/s/42d9indpjr8ac1y/TutorialEpilepsy.zip?dl=0 == Filters == === Tutorial 10: Power spectrum and frequency filters === [CODE] * '''Richard, Hossein''': Define reasonable transient durations * '''Richard, Hossein''': Update the code for the band-stop and notch filters in the same way * '''Richard, Hossein''': Frequency resolution in the "freqz" plots for high-pass filters under 0.5Hz * '''Richard, John, Hossein, Sylvain''': Address the issue of the weird PSD plots for Elekta recordings [ONLINE DOC] * '''Richard, Hossein''': Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsFilter#Filters_specifications_.5BTODO.5D|Filters specifications]] for band-stop and notch filters === Tutorial 13: Artifact cleaning with SSP === * Francois: Check the length needed to filter the recordings (after finishing #10) <<BR>>Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsSsp#SSP_Algorithm_.5BTODO.5D|SSP Algorithm]] === Tutorial 15: Import epochs === * Richard, Sylvain, John, Francois: Define recommendations for epoch lengths (after finishing #10) <<BR>>Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epoching#Epoch_length_.5BTODO.5D|Epoch length]] === Tutorial 22: Source estimation === * Francois: Update screen capture in section [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#Averaging_in_source_space|Averaging in source space]] |
| Line 92: | Line 90: |
| == Tutorial 25: Difference == [ONLINE DOC] * Validate order of the processes: Difference sources => Zscore => Low-pass filter<<BR>>Section: [[http://neuroimage.usc.edu/brainstorm/Tutorials/Difference#Difference_deviant-standard|Difference deviant-standard]] == Tutorial 26: Statistics == [CODE] * Francois: Implementation of Brainstorm-only permutation tests * Francois: Keep edge effects map (TFmask) for the stats on time-frequency maps [ONLINE DOC] * Francois: Add a section about unconstrained sources (after finishing #27) |
|
| Line 108: | Line 91: |
| * Update everything after: http://neuroimage.usc.edu/brainstorm/Tutorials/Workflows#Constrained_cortical_sources * Update: http://neuroimage.usc.edu/brainstorm/Tutorials/WorkflowGuide * Update number of pages == Tutorial 28: Scripting == * Not evaluated yet * Update number of pages |
* Add Chi2(log) ? |
| Line 117: | Line 94: |
| * Francois: Check that the script tutorial_introduction.m produces the same output as tutorials | * Francois: Remove all the wiki pages that are not used |
| Line 119: | Line 96: |
| * Francois: Remove useless images from all tutorials | |
| Line 120: | Line 98: |
<<BR>><<BR>><<BR>><<BR>><<BR>>[Additional important stuff] == Other analysis scenarios == * Francois: Update all the tutorials (100+ pages) * Francois: Remove useless images from all tutorials * Francois: Add number of pages in the tutorials * Francois: Split list in columns |
|
| Line 131: | Line 101: |
| * Richard, Sylvain: Define example dataset and precise results to obtain from them * Richard: How to assess significance from connectivity matrices * Francois: Preparation of a tutorial == Parametric experiments == * '''A = B''': Parametric t-test for constrained sources. 1. '''Sources''': Compute source maps for each trial (constrained, no normalization) 1. '''First-level statistic''': Compute a t-statistic for the source maps of all the trials A vs B. * Process2 "Test > Compute t-statistic": no absolute values, independant, equal variance. * With a high number of trials (n>30), t-values follow approximately a N(0,1) distribution. 1. '''Low-pass filter''' your evoked responses (optional). [NO! SHOULD BE DONE BEFORE, BUT WHEN ? => SPLIT ANALYSIS IN TWO: ERP OR FREQUENCY/RS] 1. '''Rectify '''the individual t-statistic (we're giving up the sign across subjects). 1. '''Project '''the individual t-statistic on a template (only when using the individual brains). 1. '''Smooth '''spatially the t-statistic maps. 1. '''Second-level statistic''': Compute a one-sampled chi-square test based on the t-statistics. * Process1: "Test > Parametric test against zero": One-sampled Chi-square test * This tests for '''|A-B|'''=0 using a Chi-square test: X = sum(|t<<HTML(<SUB>)>>i<<HTML(</SUB>)>>|^2) ~ Chi2(N<<HTML(<SUB>)>>subj<<HTML(</SUB>)>>) * Indicates when and where there is a significant effect (but not in which direction). 1. After identifying the significant effects, you may want to know which condition is stronger:<<BR>>Compute and plot power maps at the time points of interest: '''average(Ai^2^) - average(Bi^2^)''' |
* '''Richard, Sylvain''': Define example dataset and precise results to obtain from them * '''Richard, Sylvain''': How to deal with unconstrained sources?<<BR>> http://neuroimage.usc.edu/forums/showthread.php?2401 * '''Richard''': How to assess significance from connectivity matrices? * '''Richard, Hossein, Francois''': Preparation of a tutorial |
Introduction tutorials: Editing process
http://neuroimage.usc.edu/brainstorm/Tutorials
Redactors:
Francois Tadel: Montreal Neurological Institute
Elizabeth Bock: Montreal Neurological Institute
Reviewers: [current reviewing status]
Sylvain Baillet: Montreal Neurological Institute [overview 1-14, edited 20-21]
Richard Leahy: University of Southern California [validated 1-20]
John Mosher: Cleveland Clinic [edited 22 only]
Dimitrios Pantazis: Massachusetts Institute of Technology [validated 1-15]
Inverse models
Tutorial 22: Source estimation
[CODE]
John: Inverse code: Mixed head models are still not supported.
John: Explain the new (incorrect) results obtained with the epilepsy tutorial (see below)
John: Drop the option "RMS source amplitude"?
- The documentation is not informative and not encouraging at all: "RMS source amplitude: An alternative definition of SNR, but still under test and may be dropped."
- The option is not even accessible in the interface: you successively asked me to disable it for the min norm, and then made me hide the entire section "Regularization parameter" for the dipole modelling and the beamformer.
- Can I just remove it from the interface?
- Francois: Update code, tutorials and screen captures accordingly
John, Richard, Sylvain, Matti, Alex: Make the "median eigenvalue" option the default?
- John suggests to use the "median eigenvalue" option by default instead of the option "Regularize noise covariance", which as been used for many years.
In this section of the tutorials, John wrote: "Recommended option: This author (Mosher) votes for the median eigenvalue as being generally effective. The other options are useful for comparing with other software packages that generally employ similar regularization methods."
- However this modifies a lot the results: the localization results and the MN amplitudes can be very different. If this is a clear improvement, it's good to promote it. But it cannot be done randomly like this, this has to be discussed (especially with Matti and Alex) and tested.
- John: Please arrange a meeting so you can discuss this question.
John, Richard, Sylvain: Why are dSPM values 2x lower than Z-score ?
- The tutorial says "Z-normalized current density maps are also easy to interpret. They represent explicitly a "deviation from experimental baseline" as defined by the user. In contrast, dSPM indicates the deviation from the data that was used to define the noise covariance used in computing the min norm map. "
Therefore should we expect the dSPM values to deviate more from the noise recordings, than the Z-score from the pre-stim baseline? Instead of this we observe much lower values. Is there a scaling issue here?
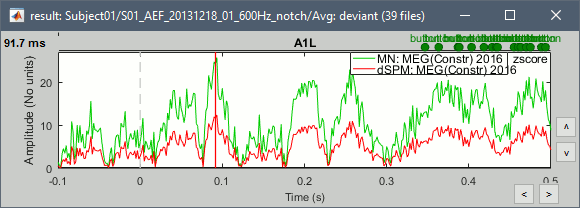
- Mixing GRAD and MAG:
John: You do not recommend processing GRAD and MAG at the same time? This is currently the default behavior in the interface...
John: Please discuss this with Matti and Alex
- Francois: Change the default + add note in Elekta tutorial if change is validated
John: Send a message to Margot Taylor: She's been asking for your beamformers for about 2yrs.
Francois: Call FieldTrip headmodels and beamformers
[ONLINE DOC]
John: Fix all the missing links
John, Richard, Sylvain: Section References
John, Richard, Sylvain: What do with Hui-Ling Beamformers?
Tutorial: Dipole scanning
John: Add the description of all the measures in this section.
Tutorial EEG/Epilepsy
John: Why sLORETA?
John: Please address the location issues with the new code:
dSPM is now localizing the spike in a much deeper spot (top=old version, bottom=new version)
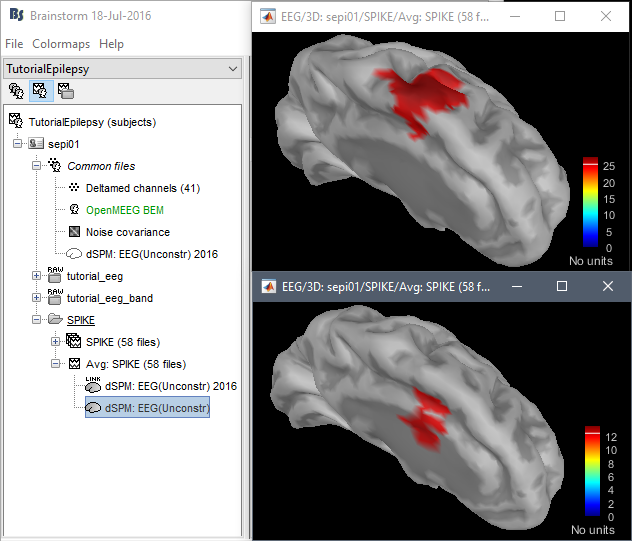
Marcel Heers wrote: "Looking at the findings from the intracranial EEG in Matthias Dümpelmann's article (figure 1 panel a) and b)) it is very likely the new sources are wrong. Additionally, the older sources are much more in agreement with Matthias' sLORETA results and with cMEM findings. Sohrabopour et al. reported as well that their IRES method found results in agreement with sources shown in the tutorial."
Imported data for testing can be downloaded here:
https://www.dropbox.com/s/42d9indpjr8ac1y/TutorialEpilepsy.zip?dl=0
Filters
Tutorial 10: Power spectrum and frequency filters
[CODE]
Richard, Hossein: Define reasonable transient durations
Richard, Hossein: Update the code for the band-stop and notch filters in the same way
Richard, Hossein: Frequency resolution in the "freqz" plots for high-pass filters under 0.5Hz
Richard, John, Hossein, Sylvain: Address the issue of the weird PSD plots for Elekta recordings
[ONLINE DOC]
Richard, Hossein: Section Filters specifications for band-stop and notch filters
Tutorial 13: Artifact cleaning with SSP
Francois: Check the length needed to filter the recordings (after finishing #10)
Section SSP Algorithm
Tutorial 15: Import epochs
Richard, Sylvain, John, Francois: Define recommendations for epoch lengths (after finishing #10)
Section Epoch length
Tutorial 22: Source estimation
Francois: Update screen capture in section Averaging in source space
Tutorial 24: Time-frequency
[CODE]
- Francois: Enable option "Hide edge effects" for Hilbert (after finishing #10)
[ONLINE DOC]
- Francois: Hilbert: Link back to the filters tutorial (after finishing #10)
Tutorial 27: Workflows
- Add Chi2(log) ?
Final steps
- Francois: Remove all the wiki pages that are not used
- Francois: Check all the links in all the pages
- Francois: Remove useless images from all tutorials
Francois: Reference on ResearchGate, Academia and Google Scholar
http://neuroimage.usc.edu/brainstorm/Tutorials/AllIntroduction
Connectivity
- Not documented at all
Richard, Sylvain: Define example dataset and precise results to obtain from them
Richard, Sylvain: How to deal with unconstrained sources?
http://neuroimage.usc.edu/forums/showthread.php?2401Richard: How to assess significance from connectivity matrices?
Richard, Hossein, Francois: Preparation of a tutorial
