|
Size: 3899
Comment:
|
Size: 4012
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 9: | Line 9: |
| Reviewers ''[current reviewing status] '' | Reviewers: ''[current reviewing status] '' |
| Line 11: | Line 11: |
| * '''Sylvain Baillet''': Montreal Neurological Institute ''[not started]'' * '''Richard Leahy''': University of Southern California ''[validated 1-9]'' * '''John Mosher''': Cleveland Clinic ''[not started]'' |
* '''Sylvain Baillet''': Montreal Neurological Institute ''[overview 1-14, edited 20-21]'' * '''Richard Leahy''': University of Southern California ''[validated 1-20]'' * '''John Mosher''': Cleveland Clinic ''[edited 22 only]'' |
| Line 16: | Line 16: |
| Expected timeline (2016) | <<BR>><<BR>> |
| Line 18: | Line 18: |
| * '''March''': Tutorials #1-#21 (interface and pre-processing) * '''April''': Tutorials #22-#24 (source estimation and time-frequency) * '''May-June''': Tutorials #25-#28 (statistics and workflows) * '''July-Sept''': Cross-validation of the pipeline and the results with MNE, FieldTrip and SPM * '''October''': Presentation during a satellite meeting at the Biomag 2016 conference * '''Nov-Dec''': Connectivity tutorial <<BR>> == Tutorial 10: Power spectrum and frequency filters == |
== Filters == === Tutorial 10: Power spectrum and frequency filters === |
| Line 30: | Line 22: |
| * Richard, John, Sylvain: Finish the evaluation of the band-pass filters used in Brainstorm * Francois: Mark with an extended event the time segments that we cannot trust * Francois: Add warning if recordings are not long enough * Francois: Check that the bandpass filter is always used with enough data (Hilbert transform, SSP) |
* Richard, Hossein: Define reasonable transient durations * Richard, Hossein: Describe the band-stop and notch filters in the same way * Richard, Hossein: Frequency resolution in the "freqz" plots for high-pass filters under 0.5Hz * Richard, John, Hossein, Sylvain: Weird PSD plots for Elekta recordings |
| Line 37: | Line 29: |
| * Richard, John, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsFilter#What_filters_to_apply.3F_.5BTODO.5D|What filters to apply?]] * Richard, John, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsFilter#Filters_specifications_.5BTODO.5D|Filters specifications]] |
* Richard, Hossein: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsFilter#Filters_specifications_.5BTODO.5D|Filters specifications]] for band-stop and notch filters |
| Line 40: | Line 31: |
| == Tutorial 13: Artifact cleaning with SSP == [CODE] [ONLINE DOC] |
=== Tutorial 13: Artifact cleaning with SSP === [CODE] |
| Line 43: | Line 34: |
| * Francois: Check the length needed to filter the recordings (after finishing tutorial #10) <<BR>>Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsSsp#SSP_Algorithm_.5BTODO.5D|SSP Algorithm]] | * Francois: Check the length needed to filter the recordings (after finishing #10) <<BR>>Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/ArtifactsSsp#SSP_Algorithm_.5BTODO.5D|SSP Algorithm]] |
| Line 45: | Line 36: |
| == Tutorial 15: Import epochs == | === Tutorial 15: Import epochs === |
| Line 50: | Line 41: |
| == Tutorial 20: Head modelling == [ONLINE DOC] |
|
| Line 53: | Line 42: |
| * John, Richard, Sylvain: In-depth review of this sensitive tutorial * John, Richard, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/AllIntroduction#Tutorials.2FHeadModel.References_.5BTODO.5D|References]] |
|
| Line 56: | Line 43: |
| == Tutorial 22: Source estimation == | == Inverse models == === Tutorial 22: Source estimation === |
| Line 59: | Line 47: |
| * John: Dipole scanning: Wrong orientations * John: Inverse code: sLORETA * John: Inverse code: NAI |
|
| Line 63: | Line 48: |
| * John, Richard, Sylvain: How to normalize unconstrained sources ? * Francois: Remove the warning messages in the interface |
* John: Explain the new (incorrect) results obtained with the epilepsy tutorial * John: Discuss and validate all the modifications with Alex and Matti * John: Cannot process GRAD and MAG at the same time ? * Francois: Make the "median" option the default? (changes a lot the range of values) |
| Line 66: | Line 53: |
| * Francois: Add note in Elekta tutorial: Process MAG and GRAD separately | |
| Line 69: | Line 57: |
| == Connectivity == * Richard, Sylvain: Define example dataset and precise results to obtain from them * Richard: Define |
* John: Fix all the missing links * John, Richard, Sylvain: Section [[http://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation#References_.5BTODO.5D|References]] * John, Richard, Sylvain: Why are dSPM values 2x lower than Z-score ? <<BR>> {{attachment:diff_zscore_dspm.gif||width="424",height="152"}} * John, Richard, Sylvain: What do with [[http://neuroimage.usc.edu/brainstorm/Tutorials/Beamformers|Hui-Ling Beamformers]]? |
| Line 73: | Line 62: |
| == Other analysis scenarios == * Francois: Update all the tutorials (100 pages) * Francois: Add number of pages in the tutorials |
=== Tutorial: Dipole scanning === * Add the description of all the measures in [[http://neuroimage.usc.edu/brainstorm/Tutorials/TutDipScan#Significance_mesures_.5BTODO.5D|this section]]. |
| Line 77: | Line 65: |
| == Phantom tutorial (for validation) == * John: Prepare recordings to distribute on the [[http://neuroimage.usc.edu/brainstorm/Tutorials/Phantom|tutorial page]] * Francois: Prepare the anatomy * Francois: Prepare an analysis script * Francois: Edit the tutorial page |
=== Tutorial EEG/Epilepsy === * Why sLORETA? * Location issues with the new code? == Tutorial 24: Time-frequency == [CODE] * Francois: Enable option "Hide edge effects" for Hilbert (after finishing #10) [ONLINE DOC] * Francois: Hilbert: Link back to the filters tutorial (after finishing #10) == Tutorial 27: Workflows == * Add Chi2(log) ? |
| Line 84: | Line 84: |
| * Check that the script tutorial_introduction.m produces the same output as the screen captures * Check all the links in all the pages * Check the number of pages for each tutorial * Reference on ResearchGate, Academia and Google Scholar: <<BR>>http://neuroimage.usc.edu/brainstorm/Tutorials/AllIntroduction |
* Francois: Remove all the wiki pages that are not used * Francois: Check all the links in all the pages * Francois: Remove useless images from all tutorials * Francois: Reference on ResearchGate, Academia and Google Scholar <<BR>>http://neuroimage.usc.edu/brainstorm/Tutorials/AllIntroduction == Connectivity == * Not documented at all * Richard, Sylvain: Define example dataset and precise results to obtain from them * Richard: How to deal with unconstrained sources?<<BR>> http://neuroimage.usc.edu/forums/showthread.php?2401 * Richard: How to assess significance from connectivity matrices? * Francois, Richard, Hossein: Preparation of a tutorial |
Introduction tutorials: Editing process
http://neuroimage.usc.edu/brainstorm/Tutorials
Redactors:
Francois Tadel: Montreal Neurological Institute
Elizabeth Bock: Montreal Neurological Institute
Reviewers: [current reviewing status]
Sylvain Baillet: Montreal Neurological Institute [overview 1-14, edited 20-21]
Richard Leahy: University of Southern California [validated 1-20]
John Mosher: Cleveland Clinic [edited 22 only]
Dimitrios Pantazis: Massachusetts Institute of Technology [validated 1-15]
Filters
Tutorial 10: Power spectrum and frequency filters
[CODE]
- Richard, Hossein: Define reasonable transient durations
- Richard, Hossein: Describe the band-stop and notch filters in the same way
- Richard, Hossein: Frequency resolution in the "freqz" plots for high-pass filters under 0.5Hz
- Richard, John, Hossein, Sylvain: Weird PSD plots for Elekta recordings
[ONLINE DOC]
Richard, Hossein: Section Filters specifications for band-stop and notch filters
Tutorial 13: Artifact cleaning with SSP
[CODE]
Francois: Check the length needed to filter the recordings (after finishing #10)
Section SSP Algorithm
Tutorial 15: Import epochs
[ONLINE DOC]
Richard, Sylvain, John, Francois: Define recommendations for epoch lengths (after finishing #10)
Section Epoch length
Inverse models
Tutorial 22: Source estimation
[CODE]
- John: Inverse code: Mixed head models
- John: Explain the new (incorrect) results obtained with the epilepsy tutorial
- John: Discuss and validate all the modifications with Alex and Matti
- John: Cannot process GRAD and MAG at the same time ?
- Francois: Make the "median" option the default? (changes a lot the range of values)
Francois: Call FieldTrip headmodels and beamformers
- Francois: Add note in Elekta tutorial: Process MAG and GRAD separately
[ONLINE DOC]
- John: Fix all the missing links
John, Richard, Sylvain: Section References
John, Richard, Sylvain: Why are dSPM values 2x lower than Z-score ?
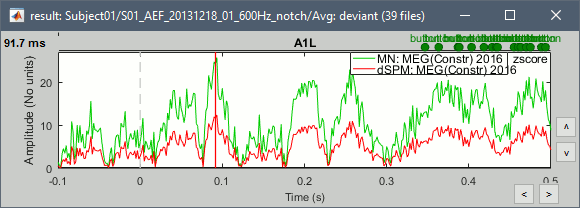
John, Richard, Sylvain: What do with Hui-Ling Beamformers?
Tutorial: Dipole scanning
Add the description of all the measures in this section.
Tutorial EEG/Epilepsy
- Why sLORETA?
- Location issues with the new code?
Tutorial 24: Time-frequency
[CODE]
- Francois: Enable option "Hide edge effects" for Hilbert (after finishing #10)
[ONLINE DOC]
- Francois: Hilbert: Link back to the filters tutorial (after finishing #10)
Tutorial 27: Workflows
- Add Chi2(log) ?
Final steps
- Francois: Remove all the wiki pages that are not used
- Francois: Check all the links in all the pages
- Francois: Remove useless images from all tutorials
Francois: Reference on ResearchGate, Academia and Google Scholar
http://neuroimage.usc.edu/brainstorm/Tutorials/AllIntroduction
Connectivity
- Not documented at all
- Richard, Sylvain: Define example dataset and precise results to obtain from them
Richard: How to deal with unconstrained sources?
http://neuroimage.usc.edu/forums/showthread.php?2401- Richard: How to assess significance from connectivity matrices?
- Francois, Richard, Hossein: Preparation of a tutorial
