|
Size: 8099
Comment:
|
Size: 14096
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| ''Authors: Takfarinas Medani ...'' | ## page was renamed from femVSbem = EEG & MEG Forward models within brainstorm: "Qualitative comparison" = ''Authors: Takfarinas Medani, ...'' |
| Line 8: | Line 10: |
| In this tutorial, we compare the head models computation using the methods available in brainstorm. This is a qualitative comparaison and not advanced study. You can find at the end of this page a list of reference that investigate deeply the differents forwad models. |
In this tutorial, we review and compare the forward models computation methods available in the brainstorm. We apply these methods in the spherical head model and in realistic models. we compare the forward solution in different scenarios. This is a qualitative comparison and not an advanced study. You can find at the end of this page a list of references that investigate deeply the differents forward models. |
| Line 20: | Line 24: |
| === The source space : === | === The source space === |
| Line 46: | Line 50: |
| In this figure, we show all of these methods and zoom to focus on the vectors. For more documentations about these lead field vectors, please refere to these pages ([[https://www.researchgate.net/publication/260603026_Biomagnetism|page1]], [[http://www.bem.fi/book/11/11x/1119x.htm|page2]]) | In this figure, we show all of these methods and zoom to focus on the vectors. |
| Line 52: | Line 56: |
| === Regarding the computation time: === * 3-shell sphere: less than 30 secondes |
For more documentations about these lead field vectors, please refer to these pages : ([[https://www.researchgate.net/publication/260603026_Biomagnetism|page1]], [[http://www.bem.fi/book/11/11x/1119x.htm|page2]]) === The computation time === Regarding the computation time: * 3-shell sphere: less than 30 seconds |
| Line 61: | Line 69: |
| From the brainstorm side, we recommend to use either openmeeg or duneuro for a realistic head model. If you have the FEM mesh*, Duneuro is faster for the same mesh resolution as the surface (OpenMeeg). | From the brainstorm side, we recommend using either OpenMeeg or duneuro for a realistic head model. If you have the FEM mesh*, Duneuro is faster for the same mesh resolution as the surface (OpenMeeg). |
| Line 68: | Line 76: |
| In this example, we will present the process of FEM mesh generation from surface model and highlight the effect of the mesh resolution. === head model === |
In this example, we will present the process of FEM mesh generation from the surface model then highlight the effect of the mesh resolution. === The head model === |
| Line 79: | Line 87: |
| MaxVol: is the maximum volume of the tetrahedral, in this example, we use the ctf coordinate system, therefore MaxVol here is a volume with units of cm3. | MaxVol: is the maximum volume of the tetrahedral, in this example, we use the CTF coordinate system, therefore MaxVol here is a volume with units of cm3. |
| Line 85: | Line 93: |
| === Leadfield computation === | === The source and sensor space === |
| Line 92: | Line 100: |
| === The forward model & lead field visualisation === | |
| Line 98: | Line 107: |
| === Lead field visualization === | |
| Line 105: | Line 113: |
| From visual checking and for different electrodes configurations(tap H for more help on the figure), we observe that there is less or difference between the V001 and V01. Also, we noticed differences in the model V1 and higher difference in the model V10 (coarse mesh). These basics observations are expected regarding the quality of the mesh. === Example on source estimation === |
From visual checking and for different electrodes configurations(tap H for more help on the figure), we observe that there is less of difference between the V001 and V01. Also, we noticed differences in the model V1 and higher difference in the model V10 (coarse mesh). These basics observations are expected regarding the quality of the mesh. === source estimation === |
| Line 112: | Line 120: |
| Repeat the process for the four head models and then we display the results on the cortex. We select a time point to highlight cortical activation. {{attachment:MNLocalisationVsMeshResolution.JPG||height="400",width="700"}} |
We repeat the process for the four head models and then we display the results on the cortex. We select a time point to highlight cortical activation. {{attachment:MNLocalisationVsMeshResolution.JPG||height="450",width="700"}} The name of each figure is displayed on the title bare. We notice that the V01 and V001 have the same range on the scaling (0-300), whereas the V1 and V10 are lower (0-200). V001 and V01 give almost the same regions and are more focal than V1 and V10. From these observations, we recommend using MaxVol = 0.1 and we set is as the default value for mesh resolution within brainstorm. === Comparaison between the different forward methods === In this section, we compute the forward model using the three available methods (3-shell, OpenMeeg, and DUNEuro). For these three methods, we use the same head model, with the same source and sensor space (as shown in the previous sections). For the Duneuro FEM, we select the head model V01 (the default value for the mesh generation). === Lead field visualisation === |
| Line 119: | Line 134: |
| Describe femVSbem here. v0.1 and V0.01 are almost the same .... the difference appears on v10 {{attachment:sourceVsMeshResolution.JPG||height="400",width="700"}} in the spherical head model same observation on the lf view {{attachment:lfview_v01andv001.JPG||height="400",width="700"}} the difference appears on the v1 and worse for v10 From these observations, we recommend using MaxVol = 0.1 and we set is as the default value for mesh resolution. == EEG forward within 3 layers head model: == In this example, we compute the head model for the ICBM head model using the openmeeg method and then we generate the fem mesh for the same subject, then we compute the lead field using the Duneuro method v = 0.1 default value. MNorm {{attachment:sourcelocalisation.JPG||height="400",width="700"}} sloreta {{attachment:sourcelocalisationloreta.JPG||height="400",width="700"}} {{attachment:sourcelocalisationloreta2.JPG||height="400",width="700"}} |
When the forward computation is completed with the three methods, we can display the lead vector. {{attachment:LeadFielOnTemplateHead.JPG||height="450",width="700"}} We can see that the LF arrows of Duneuro and 3-shell are pointing in the same direction. Whereas, for OpenMeeg, there are some outliers (big blue arrows) pointing on wrongs and random directions. When we check closely the values of the OpenMeeg LF, we notice that the vectors are pointing in the correct direction in most of the source points. The outliers have big value and could not be displayed on the same scale ([[https://github.com/brainstorm-tools/brainstorm3/issues/185#issuecomment-606886284|we are working on that]]). These points are some dipoles that are to close to the interface. The OpenMeeg BEM solution is not accurate in these positions. In the literature, similar problems are observed even with the FEM, however, this kind of instability is investigated and Duneuro offers many source models to avoid/minimize these errors. === source estimation === Following the same logic as in the previous section, we perform source localization using the three head model described below. We select the Minimum norm and we keep the default parameters. {{attachment:MNLocalisationVsMethods.JPG||height="450",width="700"}} We do the same process, but with sLoreta option : {{attachment:sourcelocalisationloreta2.JPG||height="400",width="700"}} In this, basic example, we show that all the methods point to the same area with more or less precision. As said, in this tutorial the objectify is to present how to use these tools. == MEG within a spherical model == In this part of the tutorial, we describe similar approach for the MEG. We will use the data of the [[https://neuroimage.usc.edu/brainstorm/Tutorials/PhantomCtf?highlight=(phantom)|PhantomCTF]], we recommend you to read the [[https://neuroimage.usc.edu/brainstorm/Tutorials/PhantomCtf?highlight=(phantom)|PhantomCTF tutorial]] for better understanding of this section. === The volume conductor and the source space: === With a similar approach as explained in the introduction tutorial, we can generate the BEM surfaces from the MRI of the phantom. Right-click on the subject ==> Generate BEM surfaces. This process will generate three surfaces, the inner skull, outer skull, and scalp. We will use these surfaces for the OpenMeeg BEM computation. When these surfaces are available, we can generate the FEM mesh as explained in the previous section. Right-click on the subject, then Generate FEM Mesh, Iso2mesh, MergeMesh and then keep the default options. The following figure shows the BEM and the FEM head model. {{attachment:MegHeadModelSphere.JPG||height="350",width="650"}} From the ICBM head model, we import the cortex and then align it within the phantom as shown in the figure. === The sensor model === We use a similar configuration as explained on the CTF phantom tutorial. The following figure shows the model of the sensor and the head model. {{attachment:MEGModelSensor.JPG||height="300",width="350"}} === The forward model === For the MEG, brainstorm has four methods. 1. single sphere: fitting sphere on the cortex, 20 secondes 1. Overlapping spheres: 30 secondes 1. OpenMeeg BEM: use only the inner tissue, ~10 minutes, 1. DUNEuro FEM: use only the inner mesh, ~ 15 min We use these four methods in this section. === lead field visualisation === We use a similar process to display the lead field vectors. {{attachment:MEG-DNvsOMvsOS.JPG||height="300",width="700"}} {{attachment:MEG-DNvsOM.JPG||height="300",width="700"}} These figures show the LF arrows, we can easily check the similarity on the different methods. Brainstorm recommend the use of the Overlapping spheres since it's fast and its shown that the MEG is less affected by the head geometry and the jump on the conductivity. In this section, we showed the methods available on the brainstorm. === source estimation === For each head model, we estimate the source activation of the cortex. The following figure shows the results. {{attachment:MEGsources.JPG||height="500",width="700"}} === Dipole scan === The following image shows the main results, localization, and orientation, of the fitted dipole. {{attachment:phantome_dipoleScan_duneuro_openmeeg_sSphere_OSpheresHighLight.jpg||height="500",width="700"}} === Note === Check the unit of the MEG ... a factor 1000 is noticed on the final results ... possible solution: either x 1000 the kernel, or /1000 the gain or simply check the input units of the Duneuro. == Application on the introduction tutorial == In this section, we apply the Duneuro computation for the dataset sample of the brainstorm tutorial. === Head moel generation, forward model and source estimation === Follow the steps explained on the[[https://neuroimage.usc.edu/brainstorm/Duneuro?highlight=(duneuro)|FEM tutorial]] the follows the same step to use the Duneuro model to compute the source as explained in [[https://neuroimage.usc.edu/brainstorm/Tutorials/SourceEstimation?highlight=(source)|this tutorial]]. {{attachment:introSubjectFem.JPG||height="300",width="700"}} Here is a view of the lead vectors for the two models. {{attachment:introSubjectLeadFieldVector.JPG||height="300",width="700"}} {{attachment:introSubjectLeadFieldVectorZoom.JPG||height="300",width="700"}} === Comparaison of the source localisation === We compute the source using the MN with a constrained source. In this figure, we show the obtained results {{attachment:IntroTutoSourceLocalisationVew1.JPG||height="500",width="700"}} {{attachment:IntroTutoSourceLocalisationVew2.JPG||height="250",width="700"}} These figures show similare activated regions (Gduneuro/100 > computeSource). |
EEG & MEG Forward models within brainstorm: "Qualitative comparison"
Authors: Takfarinas Medani, ...
[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE]
Contents
Introduction
In this tutorial, we review and compare the forward models computation methods available in the brainstorm.
We apply these methods in the spherical head model and in realistic models. we compare the forward solution in different scenarios.
This is a qualitative comparison and not an advanced study. You can find at the end of this page a list of references that investigate deeply the differents forward models.
At this time, the FEM implementation is available for the EEG and the MEG computation. We will integrate and test soon the FEM for sEEG and ECOG.
EEG within a spherical model
The volume conductor or the head model:
In this part we used the spherical head model distributed by the duneuro team (sphere). The following figure shows on the left the surface model and on right the tetrahedral mesh.
The model has three layers, the brain (inner skull), the outer skull and the scalp.
The source space
For the source space or dipole position, we will use a realistic cortex distributed with the ICBM default subject of brainstorm.
The sensor model
Regarding the electrode's positions, we use the same position defined in this file. The total number of electrodes is 200 regularly distributed on the outer layer.
The forward model
Now, we have all the components of the model, we can start the process to compute the head model. For the EEG, brainstorm offers three methods. We perform these three computations on this model.
- 3-shell sphere: best-fitting sphere based on the scalp, then OK.
OpenMeeg BEM: use the conductivity 0.33, 0.004 and 0.33, and keep the default options.
- DUNEuro FEM: use the default option with the same conductivity value as the previous method.
Qualitative comparison of the methods
In this tutorial, we use the display of the lead field vector to compare visually the performances of the three methods.
Right-click on the head model, and then select 'View lead field vector'. You can select more than the head model in order to overlay the vectors.
In this figure, we show all of these methods and zoom to focus on the vectors.
We can easily see that the three vectors are pointing in the same direction and have the same length.
For more documentations about these lead field vectors, please refer to these pages : (page1, page2)
The computation time
Regarding the computation time:
- 3-shell sphere: less than 30 seconds
OpenMeeg BEM: ~ 12 minutes.
- DUNEuro FEM: ~ 2 minutes.
This example has 15000 dipoles (x3) and 200 electrodes.
There are many studies and publications that compare these methods and highlights the advantages and weakness of each method.
From the brainstorm side, we recommend using either OpenMeeg or duneuro for a realistic head model. If you have the FEM mesh*, Duneuro is faster for the same mesh resolution as the surface (OpenMeeg).
Duneuro can be used for a more realistic model (more than 3 layers with the complex shape).
*You can generate the FEM if you have the surfaces or the MRI, this process is explained in this tutorial.
EEG within brainstorm template (3 layers)
In this example, we will present the process of FEM mesh generation from the surface model then highlight the effect of the mesh resolution.
The head model
We will use the default subject, then right-click and generate FEM mesh, and follow the same steps as explained in this section.
We repeated this process four times, and hen we generate four FEM head models with four different values of the 'Max tetrahedral volume'.
Here is a view of the obtained mesh with MaxVol = 10, 1, 0.1 and 0.01 (clockwise order).
Let's give the name V10, V1, V01, and V001 respectively for these head models.
MaxVol: is the maximum volume of the tetrahedral, in this example, we use the CTF coordinate system, therefore MaxVol here is a volume with units of cm3.
The FEM generation time is quite fast, it varies from 15 secondes (V10) to less than 2 minutes (V001).
The source and sensor space
We use a protocol with EEG data, you can reproduce with any protocol with available channels position and EEG recording, this is not important in this tutorial.
For the source space, we use the default cortex with 15000 dipoles. The number of channels is 31, regularly distributed on the scalp surface, the model is shown on this figure.
The forward model & lead field visualisation
Switch to the "functional data" view, right click on the subject => Compute Head model => Duneuro FEM ==> keep the defaults parameters and set the conductivity to 0.33, 0.04 and 0.33.
Repeat the same process for four head models.
The computation time in these models is not expensive, it varies from 1 minute (V10) to 6 minutes(V001).
As in the previous example, we will display the lead field vectors for these head models.
With the FEM, it's known that increasing the mesh increases the accuracy of the solution. Therefore we will use the V001 as a reference solution and we compare the other model to this reference.
From visual checking and for different electrodes configurations(tap H for more help on the figure), we observe that there is less of difference between the V001 and V01. Also, we noticed differences in the model V1 and higher difference in the model V10 (coarse mesh). These basics observations are expected regarding the quality of the mesh.
source estimation
In this section, we perform a source estimation using the previous four head models.
Right-click on the head model ==> compute source [2018] and keep all the parameters as they are.
We repeat the process for the four head models and then we display the results on the cortex. We select a time point to highlight cortical activation.
The name of each figure is displayed on the title bare. We notice that the V01 and V001 have the same range on the scaling (0-300), whereas the V1 and V10 are lower (0-200). V001 and V01 give almost the same regions and are more focal than V1 and V10.
From these observations, we recommend using MaxVol = 0.1 and we set is as the default value for mesh resolution within brainstorm.
Comparaison between the different forward methods
In this section, we compute the forward model using the three available methods (3-shell, OpenMeeg, and DUNEuro). For these three methods, we use the same head model, with the same source and sensor space (as shown in the previous sections). For the Duneuro FEM, we select the head model V01 (the default value for the mesh generation).
Lead field visualisation
the BEM, FEM forward computation in basic model (realistic head model 3 layer) and also within a spherical head model with an analytical solution.
When the forward computation is completed with the three methods, we can display the lead vector.
We can see that the LF arrows of Duneuro and 3-shell are pointing in the same direction. Whereas, for OpenMeeg, there are some outliers (big blue arrows) pointing on wrongs and random directions.
When we check closely the values of the OpenMeeg LF, we notice that the vectors are pointing in the correct direction in most of the source points. The outliers have big value and could not be displayed on the same scale (we are working on that).
These points are some dipoles that are to close to the interface. The OpenMeeg BEM solution is not accurate in these positions. In the literature, similar problems are observed even with the FEM, however, this kind of instability is investigated and Duneuro offers many source models to avoid/minimize these errors.
source estimation
Following the same logic as in the previous section, we perform source localization using the three head model described below.
We select the Minimum norm and we keep the default parameters.
We do the same process, but with sLoreta option :
In this, basic example, we show that all the methods point to the same area with more or less precision.
As said, in this tutorial the objectify is to present how to use these tools.
MEG within a spherical model
In this part of the tutorial, we describe similar approach for the MEG. We will use the data of the PhantomCTF, we recommend you to read the PhantomCTF tutorial for better understanding of this section.
The volume conductor and the source space:
With a similar approach as explained in the introduction tutorial, we can generate the BEM surfaces from the MRI of the phantom. Right-click on the subject ==> Generate BEM surfaces.
This process will generate three surfaces, the inner skull, outer skull, and scalp. We will use these surfaces for the OpenMeeg BEM computation. When these surfaces are available, we can generate the FEM mesh as explained in the previous section. Right-click on the subject, then Generate FEM Mesh, Iso2mesh, MergeMesh and then keep the default options. The following figure shows the BEM and the FEM head model.
From the ICBM head model, we import the cortex and then align it within the phantom as shown in the figure.
The sensor model
We use a similar configuration as explained on the CTF phantom tutorial. The following figure shows the model of the sensor and the head model.
The forward model
For the MEG, brainstorm has four methods.
- single sphere: fitting sphere on the cortex, 20 secondes
- Overlapping spheres: 30 secondes
OpenMeeg BEM: use only the inner tissue, ~10 minutes,
- DUNEuro FEM: use only the inner mesh, ~ 15 min
We use these four methods in this section.
lead field visualisation
We use a similar process to display the lead field vectors.
These figures show the LF arrows, we can easily check the similarity on the different methods.
Brainstorm recommend the use of the Overlapping spheres since it's fast and its shown that the MEG is less affected by the head geometry and the jump on the conductivity.
In this section, we showed the methods available on the brainstorm.
source estimation
For each head model, we estimate the source activation of the cortex. The following figure shows the results.
Dipole scan
The following image shows the main results, localization, and orientation, of the fitted dipole.
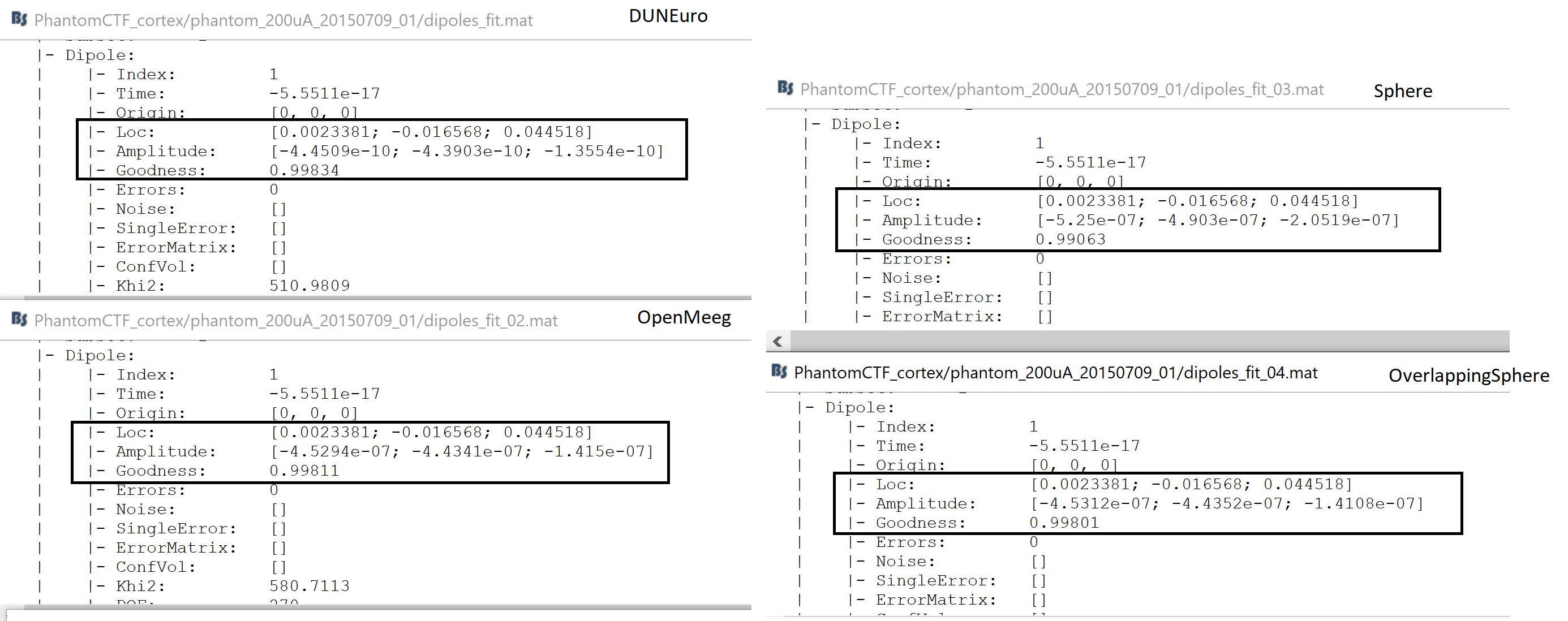
Note
Check the unit of the MEG ... a factor 1000 is noticed on the final results ... possible solution: either x 1000 the kernel, or /1000 the gain or simply check the input units of the Duneuro.
Application on the introduction tutorial
In this section, we apply the Duneuro computation for the dataset sample of the brainstorm tutorial.
Head moel generation, forward model and source estimation
Follow the steps explained on theFEM tutorial the follows the same step to use the Duneuro model to compute the source as explained in this tutorial.
Here is a view of the lead vectors for the two models.
Comparaison of the source localisation
We compute the source using the MN with a constrained source. In this figure, we show the obtained results
These figures show similare activated regions (Gduneuro/100 > computeSource).
