|
Size: 12967
Comment:
|
Size: 14731
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| You can use the free !BrainSuite and SVReg software package to extract segmented brain surfaces from a T1-weighted MRI image. Surface extraction and segmentation can either be automatic or manual--this tutorial will step you through the automated process (more information on manual tweaks can be found in [[http://brainsuite.loni.ucla.edu/processing/|BrainSuite's documentation]]). | ''Authors: Andrew Krause, Francois Tadel, Anand Joshi, David Shattuck'' |
| Line 4: | Line 4: |
| For this tutorial, we will use the 01.nii file from the CTF tutorials. If you do not already have those files on your computer, you can go to the [brainstorm3_register/download.php|Download page]] and get the sample_ctf.zip file. After extracting the zip file, the MRI image we will be using can be found in the Anatomy folder. | You can use the free BrainSuite and SVReg software package to extract segmented brain surfaces from a T1-weighted MRI image. Surface extraction and segmentation can either be automatic or manual--this tutorial will step you through the automated process (more information on manual tweaks can be found in [[http://brainsuite.org/processing/|BrainSuite's documentation]]). |
| Line 6: | Line 6: |
| Unlike !BrainStorm, Freesurfer, and !BrainVisa, !BrainSuite does not use a database structure for storing subject information and scans. Instead, by default it stores its output in the same folder as the input MRI scan. Thus, it is often best to have each subject's structural scan in its own folder. | For this tutorial, we will use the 01.nii file from the auditory tutorials. If you do not already have these files on your computer, you can go to the [[http://neuroimage.usc.edu/bst/download.php|Download page]] and get the sample_auditory.zip file. After extracting the zip file, the MRI image we will be using can be found in the Anatomy folder. |
| Line 8: | Line 8: |
| To extract head and cortex meshes from a T1 MRI, you can also try to use: [[Tutorials/SegBrainVisa|BrainVisa]] or [[Tutorials/SegFreeSurfer|FreeSurfer]]. | '''[TODO]''' Update documentation to process the [[http://neuroimage.usc.edu/bst/getupdate.php?s=auditory_tutorial_T1&u=mne|original MRI]]. Unlike Brainstorm, Freesurfer and BrainVisa, BrainSuite does not use a database structure for storing subject information and scans. Instead, by default it stores its output in the same folder as the input MRI scan. Thus, it is often best to have each subject's structural scan in its own folder. <<TableOfContents(2,2)>> |
| Line 11: | Line 15: |
| 1. Download the latest version of !BrainSuite from http://www.loni.ucla.edu/Software/BrainSuite13. 1. Install it on your computer by following the instructions in [[http://brainsuite.loni.ucla.edu/quickstart/installation/|BrainSuite's quick start installation guide]]. Note that you will be using Surface/Volume Registration (SVReg) to do the surface segmentation, so you will need to follow the instructions in the pink box on the top of the page to download a compatible MATLAB Compiler Runtime. You may ignore any instructions about "Setting up BDP" if you are not planning on using !BrainSuite for diffusion imaging. 1. Start !BrainSuite (if it is not already open following installation). |
1. Download the latest version of BrainSuite from http://www.brainsuite.org/download. 1. Install it on your computer by following the instructions in [[http://brainsuite.bmap.ucla.edu/quickstart/installation/|BrainSuite's quick start installation guide]]. 1. Note that you will be using Surface/Volume Registration (SVReg) to do the surface segmentation, so you need to install a compatible [[http://www.mathworks.com/products/compiler/mcr|MATLAB Compiler Runtime]](2015b for BrainSuite 16a). You may ignore any instructions about "Setting up BDP" if you are not planning on using BrainSuite for diffusion imaging. 1. Start BrainSuite (if it is not already open following installation). |
| Line 16: | Line 21: |
| 1. Open the T1-weighted scan in !BrainSuite by going to File > Open Volume... and navigating to the CTF tutorial's 01.nii scan.<<BR>> {{attachment:openvolume.png}} 1. Check that the scan is in the correct orientation. Some of the modules !BrainSuite and SVReg run while processing a scan assume that the scan is in LPI coordinates. Practically, this means the screen should look like below when you open it.<<BR>> {{attachment:correctorientationsmall.png}} 1. Open the Cortical Surface Extraction sequence dialog by clicking on Cortex > Cortical Surface Extraction Sequence.<<BR>> {{attachment:opencorticalsurface.png}} 1. Check the box next to "Register and label brain".<<BR>> {{attachment:csedialog.png}} 1. At the bottom of the dialog window, check that the filename prefix is "01" and that the working directory is the Anatomy folder of the CTF tutorial files (or, if you moved the scan to its own folder, that folder). 1. Click "Run All". The full surface creation and segmentation process should take about 1 - 1.5 hours, depending on your machine.<<BR>> {{attachment:completedsegmentationsmall.png}} |
1. Open the T1-weighted scan in BrainSuite by going to '''File > Open Volume'''... and navigating to the auditory tutorial's T1.nii scan.<<BR>><<BR>> {{attachment:brainsuite1small.png}} 1. Check that the scan is in the correct orientation. Some of the modules BrainSuite and SVReg run while processing a scan assume that the scan is in LPI coordinates. Practically, this means the screen should look like below when you open it.<<BR>><<BR>> {{attachment:brainsuite2small.png}} 1. Clicking on menu '''Cortex > Cortical Surface Extraction Sequence'''. <<BR>><<BR>> 1. Check the box next to "'''Register and label brain'''".<<BR>><<BR>> {{attachment:brainsuite14small.png}} 1. BrainSuite has two atlases, '''BrainSuiteAtlas1''' and '''BCI-DNI_brain_atlas'''. If you are using '''BrainSuiteAtlas1 '''here, then you should use '''Colin27_BrainSuite_2016 '''or '''ICBM152_BrainSuite_2016''' as the default anatomy.''' '''If you are using the '''BCI-DNI_brain_atlas '''here, then you should use '''BCI-DNI_BrainSuite_2016 '''as the default anatomy in BrainStorm. 1. At the bottom of the dialog window, check that the filename prefix is "T1" and that the working directory is the Anatomy folder of the CTF tutorial files (or, if you moved the scan to its own folder, that folder). 1. Click "Run All". The full surface creation and segmentation process should take about 0.5 - 1.5 hours, depending on your machine.<<BR>><<BR>> {{attachment:brainsuite13small_v2.png}} |
| Line 26: | Line 32: |
| 1. Right-click on the subject > Import !BrainSuite folder...<<BR>> {{attachment:importbrainsuitecropped.png}} 1. Select the top folder of your subject (i.e. the "Working Directory" from !BrainSuite) 1. Then you're prompted for the number of vertices you want in the final cortex surface. This will by extension define the number of dipoles to estimate during the source estimation process. By default we set this value to 15000 for the entire brain (it means 7500 for each hemisphere). <<BR>> {{attachment:nvertices.png}} 1. The MRI Viewer appears, and a help window asks you to validate the orientation of the MRI and to define the 6 fiducial points. If something doesn't look right at this step, for instance if the MRI is not presented with a correct orientation, you should stop this automatic import process and follow the manual instructions in the basic tutorial pages.<<BR>> {{attachment:fiducialssmall.png}} 1. Place the six fiducials. If you need help, refer to this page: CoordinateSystems |
1. Right-click on the subject > Import anatomy folder...<<BR>><<BR>> {{attachment:import1.gif||height="254",width="212"}} 1. Select the file format "BrainSuite folder" and select the top folder of your subject (i.e. the "Working Directory" from BrainSuite) 1. Then you're prompted for the number of vertices you want in the final cortex surface. This will by extension define the number of dipoles to estimate during the source estimation process. By default we set this value to 15000 for the entire brain (it means 7500 for each hemisphere). <<BR>><<BR>> {{attachment:nvertices.png}} 1. The MRI Viewer appears, and a help window asks you to validate the orientation of the MRI and to define the 6 fiducial points. If something doesn't look right at this step, for instance if the MRI is not presented with a correct orientation, you should stop this automatic import process and follow the manual instructions in the basic tutorial pages.<<BR>><<BR>> {{attachment:fiducialssmall.png||height="400",width="415"}} 1. Click on "'''Compute MNI transformation'''". 1. Place the three fiducials NAS/LPA/RPA. If you need help, refer to this page: [[CoordinateSystems]] |
| Line 32: | Line 39: |
| 1. The files that are imported from the !BrainSuite folder are the following: * '''01.nii''' (original T1 MRI volume) * '''01.scalp.dfs''' (head surface) * '''01.inner_skull.dfs''' (inner skull surface) * '''01.outer_skull.dfs''' (outer skull surface) * '''01.left.pial.cortex.svreg.dfs''' (grey/csf interface, left hemisphere) * '''01.right.pial.cortex.svreg.dfs''' (grey/csf interface, right hemisphere) * '''01.left.inner.cortex.svreg.dfs''' (white matter, left hemisphere) * '''01.right.inner.cortex.svreg.dfs''' (white matter, right hemisphere) |
1. The files that are imported from the BrainSuite folder are the following: * '''T1.nii''' (original T1 MRI volume) * '''T1.scalp.dfs''' (head surface) * '''T1.inner_skull.dfs''' (inner skull surface) * '''T1.outer_skull.dfs''' (outer skull surface) * '''T1.left.pial.cortex.svreg.dfs''' (grey/csf interface, left hemisphere) * '''T1.right.pial.cortex.svreg.dfs''' (grey/csf interface, right hemisphere) * '''T1.left.inner.cortex.svreg.dfs''' (white matter, left hemisphere) * '''T1.right.inner.cortex.svreg.dfs''' (white matter, right hemisphere) |
| Line 42: | Line 49: |
| 1. The successive steps that are performed automatically by Brainstorm: | 1. The successive steps that are automatically performed by Brainstorm: |
| Line 44: | Line 51: |
| * Read the anatomical label information from pial and white matter surfaces | * Read the anatomical label information and SVReg atlases from pial and white matter surfaces |
| Line 46: | Line 53: |
| * Merge left and right hemipsheres for the two cortical surface types: white matter and cortex envelope | * Merge left and right hemispheres for the two cortical surface types: white matter and cortex envelope |
| Line 48: | Line 55: |
| * Delete all the uncessary surfaces 1. The files you can see in the database explorer at the end:<<BR>> {{attachment:brainsuitefilescropped.png}} |
* Delete all the unnecessary surfaces 1. The files you can see in the database explorer at the end:<<BR>><<BR>> {{attachment:brainsuitefilescropped.png}} |
| Line 54: | Line 61: |
| * '''cortex_270604V''': High-resolution cortical surface that was generated by !BrainSuite. | * '''cortex_270604V''': High-resolution cortical surface that was generated by BrainSuite. |
| Line 56: | Line 63: |
| 1. A figure is automatically shown at the end of the process, to check visually that the low-resolution cortex and head surfaces were properly imported. If it doesn't look like the following picture, do not go any further in your source analysis, fix the anatomy first.<<BR>> {{attachment:importresultssmall.png}} | 1. A figure is automatically shown at the end of the process, to check visually that the low-resolution cortex and head surfaces were properly imported. If it doesn't look like the following picture, do not go any further in your source analysis, fix the anatomy first.<<BR>><<BR>> {{attachment:importresultssmall.png||height="179",width="227"}} |
| Line 64: | Line 71: |
| {{attachment:alignmrisurfacesmall.png}} | . {{attachment:alignmrisurfacesmall.png||height="273",width="226"}} |
| Line 67: | Line 74: |
| It is critical to get a good cortex surface for source estimation. If the final cortex surface looks bad, it means that something didn't work well somewhere along the !BrainSuite cortical surface extraction sequence. This tutorial is not meant to guide you though troubleshooting !BrainSuite results, but most errors in surface generation and segmentation occur at the first step, skull-stripping. If you are ending up with surfaces that include non-brain regions, or are missing large chunks of cortex, this step is where the error occured. | It is critical to get a good cortex surface for source estimation. If the final cortex surface looks bad, it means that something didn't work well somewhere along the BrainSuite cortical surface extraction sequence. This tutorial is not meant to guide you though troubleshooting BrainSuite results, but most errors in surface generation and segmentation occur at the first step, skull-stripping. If you are ending up with surfaces that include non-brain regions, or are missing large chunks of cortex, this step is where the error occurred. |
| Line 69: | Line 76: |
| To fix skull-stripping issues, re-open the original T1 scan in !BrainSuite. Open the Cortical Surface Extraction Sequence and check the box next to "Register and label brain" as you did above, but this time uncheck "Skull stripping". Next click on Cortex > Skull Stripping (BSE) to open the skull stripping dialog box, as this will let you step back and forth until you get a result that works. Common paramter settings that work are: | To fix skull-stripping issues, re-open the original T1 scan in BrainSuite. Open the Cortical Surface Extraction Sequence and check the box next to "Register and label brain" as you did above, but this time uncheck "Skull stripping". Next click on Cortex > Skull Stripping (BSE) to open the skull stripping dialog box, as this will let you step back and forth until you get a result that works. Common parameter settings that work are: |
| Line 73: | Line 80: |
| * Erosion Size: 1, 2 | |
| Line 74: | Line 82: |
| In general, increasing the edge constant will exclude more areas of the brain (decreasing will include more areas), while increasing the diffusion constant will include more areas. Set the parameters as you want, then click "Step >" until it says "finished skullstripping" above the progress bar. If the results look good, then click "Run All" on the Cortical Surface Extraction Sequence dialog box to run the rest of the segmentation process; if the results do not look good, click "< Back" on the Skull Stripping Tool dialog box until the progress bar returns to 0% and try a different set of parameters. More information can be found in [[http://brainsuite.loni.ucla.edu/processing/surfaceextraction/bse/tuning-the-bse-parameters/|BrainSuite's documentation]]. | In general, increasing the edge constant will exclude more areas of the brain (decreasing will include more areas), while increasing the diffusion constant will include more areas. Set the parameters as you want, then click "Step >" until it says "finished skullstripping" above the progress bar. If the results look good, then click "Run All" on the Cortical Surface Extraction Sequence dialog box to run the rest of the segmentation process; if the results do not look good, click "< Back" on the Skull Stripping Tool dialog box until the progress bar returns to 0% and try a different set of parameters. More information can be found in [[http://brainsuite.bmap.ucla.edu/processing/surfaceextraction/bse/tuning-the-bse-parameters/|BrainSuite's documentation]]. |
| Line 76: | Line 84: |
| If after following those instructions you still don't manage to get good surfaces, you can try to run the automatic MRI segmentation from [[Tutorials/SegBrainVisa|BrainVISA]] or [[Tutorials/LabelFreeSurfer|FreeSurfer]]. | If after following these instructions you still don't manage to get good surfaces, you can try to run the automatic MRI segmentation from [[Tutorials/SegBrainVisa|BrainVISA]] or [[Tutorials/LabelFreeSurfer|FreeSurfer]]. |
| Line 81: | Line 89: |
| The head surface is important mostly for the alignment of the MEG sensors and the MRI. If you digitized the head shape with a Polhemus device, you can align automatically the head surface (hence the MRI) with the MEG sensors (in the same referential as the Polhemus points). The quality of this automatic registrations depends on the quality of both surfaces: the Polhemus head shape (green points) and the head surface from the MRI (grey surface). If you placed lots of points on the nose but your head surface doesn't have a nose, those points are not going to help. Except for that, a nice head shape is mainly useful for producing nicer figures. | The head surface is important mostly for the alignment of the MEG sensors and the MRI. If you digitized the head shape with a Polhemus device, you can automatically align the head surface (hence the MRI) with the MEG sensors (in the same referential as the Polhemus points). The quality of this automatic registrations depends on the quality of both surfaces: the Polhemus head shape (green points) and the head surface from the MRI (grey surface). If you placed lots of points on the nose but your head surface doesn't have a nose, these points are not going to help. Except for that, a nice head shape is mainly useful for producing nicer figures. |
| Line 91: | Line 99: |
| You can also try to import manually the original !BrainSuite head surface. Right click on the subject > Import surface > 01.scalp.dfs. If the surface has holes, you can right-click on the head surface > Fill holes. Note that !BrainSuite's head and skull files have too many vertices for analysis, so you may have to decrease the surface resolution by right-clicking the head file > Less vertices.... The automated folder importation decreases the number of vertices to 1082. | You can also try to import manually the original BrainSuite head surface. Right click on the subject > Import surface > 01.scalp.dfs. If the surface has holes, you can right-click on the head surface > Fill holes. Note that BrainSuite's head and skull files have too many vertices for analysis, so you may have to decrease the surface resolution by right-clicking the head file > Less vertices.... The automated folder importation decreases the number of vertices to 1082. |
| Line 94: | Line 102: |
| Checking the box next to "Register and label brain" when running !BrainSuite's Cortical Surface Extraction sequence causes !BrainSuite to run a companion program called SVReg that implements an automatic parcellation of the cortical surface into anatomical regions. More information on this program can be found here:http://brainsuite.loni.ucla.edu/processing/svreg/ | Checking the box next to "Register and label brain" when running BrainSuite's Cortical Surface Extraction sequence causes BrainSuite to run a companion program called SVReg that implements an automatic parcellation of the cortical surface into anatomical regions. More information on this program can be found here:http://brainsuite.bmap.ucla.edu/processing/svreg/ |
| Line 96: | Line 104: |
| SVReg's labels are imported into !BrainStorm as scouts (cortical regions of interest), and saved directly in the surface files. To check where they are saved: right-click on the low-resolution cortex file > File > View file contents. You can see that there are two "Atlas" structures available, one that has Name='User scouts' and the other with Name='SVReg'. | SVReg's labels are imported into BrainStorm as scouts (cortical regions of interest), and saved directly in the surface files. To check where they are saved: right-click on the low-resolution cortex file > File > View file contents. You can see that there are two "Atlas" structures available, one that has Name='User scouts' and the other with Name='SVReg'. |
| Line 100: | Line 108: |
| {{attachment:svregscoutssmall.png}} | {{attachment:svregscoutssmall.png||height="252",width="621"}} |
| Line 102: | Line 110: |
| == Feedback == <<EmbedContent(http://neuroimage.usc.edu/brainstorm3_register/get_feedback.php?Tutorials/SegBrainSuite)>> |
==== Troubleshooting: You do not see the atlas SVReg ==== If you get some error messages in the Matlab command window during the segmention ("Could not find XML label description file brainsuite_labeldescription.xml"), you are using a version of BrainSuite that does not save the file brainsuite_labeldescription.xml in the segmentation folder. Workaround: Copy manually the file brainsuite_labeldescription.xml to the segmentation folder. == Template registration for group analysis == BrainSuite provides an accurate registration method to the Brainstorm anatomy templates (Colin27, ICBM152). The registration procedure works in the same way as the FreeSurfer-based registration illustrated in this tutorial: [[Tutorials/CoregisterSubjects|Group studies: Subject coregistration]]. To use this registration, you need to use a template anatomy processed with BrainSuite. <<BR>>Right-click on ''Default anatomy'' > Use template > '''Colin27_BrainSuite_2016''' or '''ICBM152_BrainSuite_2016'''. . {{attachment:coreg.gif||height="331",width="554"}} <<EmbedContent(http://neuroimage.usc.edu/bst/get_feedback.php?Tutorials/SegBrainSuite)>> |
T1-MRI Segmentation with BrainSuite
Authors: Andrew Krause, Francois Tadel, Anand Joshi, David Shattuck
You can use the free BrainSuite and SVReg software package to extract segmented brain surfaces from a T1-weighted MRI image. Surface extraction and segmentation can either be automatic or manual--this tutorial will step you through the automated process (more information on manual tweaks can be found in BrainSuite's documentation).
For this tutorial, we will use the 01.nii file from the auditory tutorials. If you do not already have these files on your computer, you can go to the Download page and get the sample_auditory.zip file. After extracting the zip file, the MRI image we will be using can be found in the Anatomy folder.
[TODO] Update documentation to process the original MRI.
Unlike Brainstorm, Freesurfer and BrainVisa, BrainSuite does not use a database structure for storing subject information and scans. Instead, by default it stores its output in the same folder as the input MRI scan. Thus, it is often best to have each subject's structural scan in its own folder.
Contents
Installation
Download the latest version of BrainSuite from http://www.brainsuite.org/download.
Install it on your computer by following the instructions in BrainSuite's quick start installation guide.
Note that you will be using Surface/Volume Registration (SVReg) to do the surface segmentation, so you need to install a compatible MATLAB Compiler Runtime(2015b for BrainSuite 16a). You may ignore any instructions about "Setting up BDP" if you are not planning on using BrainSuite for diffusion imaging.
Start BrainSuite (if it is not already open following installation).
Running BrainSuite
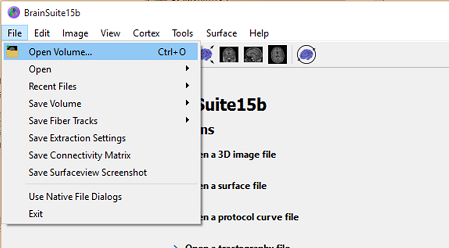
Open the T1-weighted scan in BrainSuite by going to File > Open Volume... and navigating to the auditory tutorial's T1.nii scan.
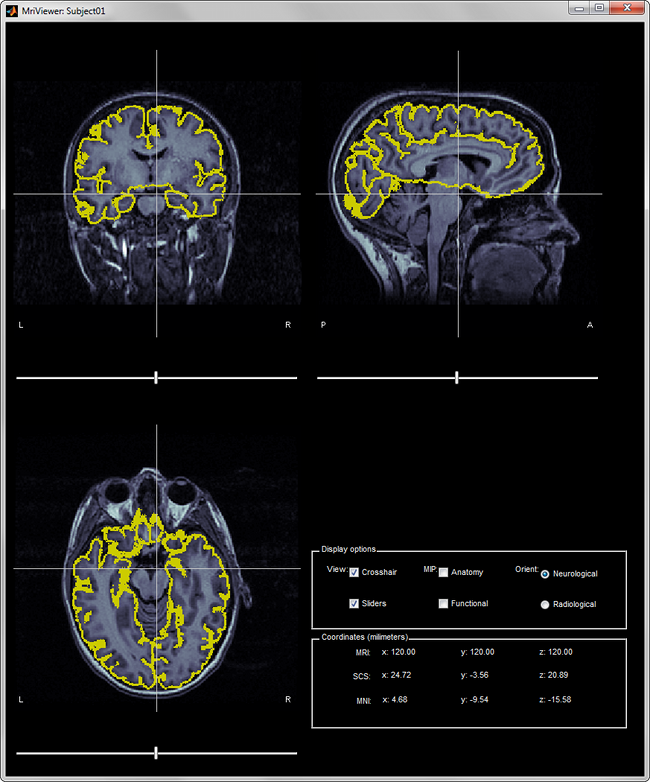
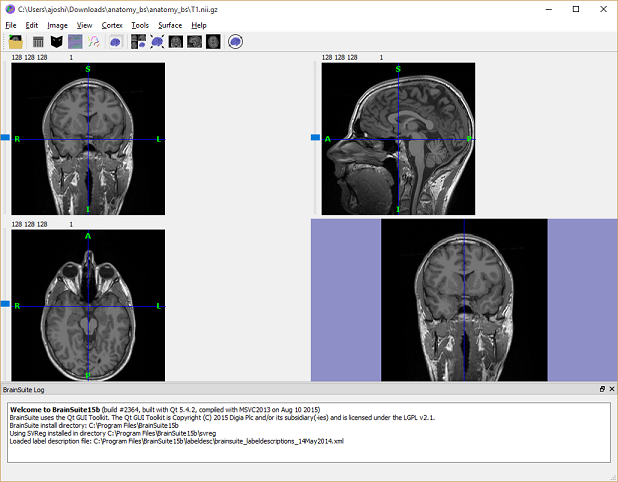
Check that the scan is in the correct orientation. Some of the modules BrainSuite and SVReg run while processing a scan assume that the scan is in LPI coordinates. Practically, this means the screen should look like below when you open it.
Clicking on menu Cortex > Cortical Surface Extraction Sequence.
Check the box next to "Register and label brain".
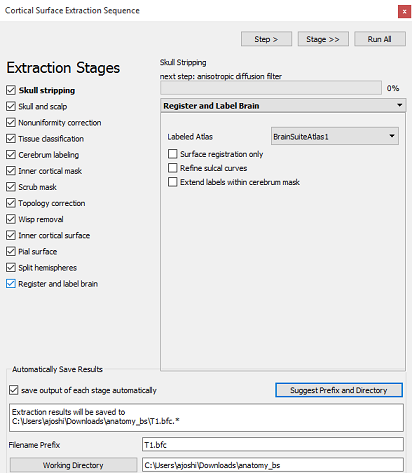
BrainSuite has two atlases, BrainSuiteAtlas1 and BCI-DNI_brain_atlas. If you are using BrainSuiteAtlas1 here, then you should use Colin27_BrainSuite_2016 or ICBM152_BrainSuite_2016 as the default anatomy. If you are using the BCI-DNI_brain_atlas here, then you should use BCI-DNI_BrainSuite_2016 as the default anatomy in BrainStorm.
- At the bottom of the dialog window, check that the filename prefix is "T1" and that the working directory is the Anatomy folder of the CTF tutorial files (or, if you moved the scan to its own folder, that folder).
Click "Run All". The full surface creation and segmentation process should take about 0.5 - 1.5 hours, depending on your machine.
Importing the results in Brainstorm
- Switch to the anatomy side of the database explorer
- Create a new subject, set the default anatomy option to "No, use individual anatomy"
Right-click on the subject > Import anatomy folder...
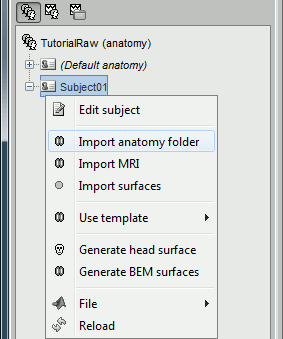
Select the file format "BrainSuite folder" and select the top folder of your subject (i.e. the "Working Directory" from BrainSuite)
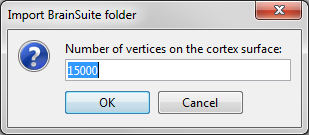
Then you're prompted for the number of vertices you want in the final cortex surface. This will by extension define the number of dipoles to estimate during the source estimation process. By default we set this value to 15000 for the entire brain (it means 7500 for each hemisphere).
The MRI Viewer appears, and a help window asks you to validate the orientation of the MRI and to define the 6 fiducial points. If something doesn't look right at this step, for instance if the MRI is not presented with a correct orientation, you should stop this automatic import process and follow the manual instructions in the basic tutorial pages.
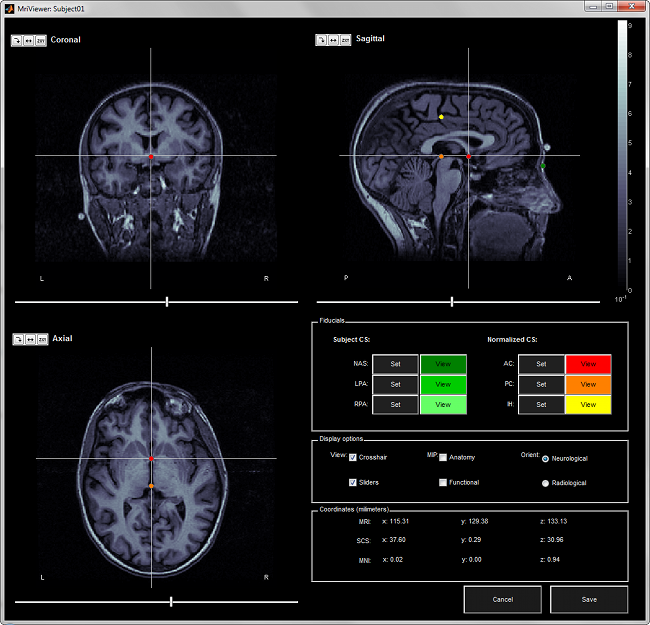
Click on "Compute MNI transformation".
Place the three fiducials NAS/LPA/RPA. If you need help, refer to this page: CoordinateSystems
- Click on Save to keep your modifications, and the automatic import will go on.
The files that are imported from the BrainSuite folder are the following:
T1.nii (original T1 MRI volume)
T1.scalp.dfs (head surface)
T1.inner_skull.dfs (inner skull surface)
T1.outer_skull.dfs (outer skull surface)
T1.left.pial.cortex.svreg.dfs (grey/csf interface, left hemisphere)
T1.right.pial.cortex.svreg.dfs (grey/csf interface, right hemisphere)
T1.left.inner.cortex.svreg.dfs (white matter, left hemisphere)
T1.right.inner.cortex.svreg.dfs (white matter, right hemisphere)
brainsuite_labeldescription.xml (not imported, but required for properly labelling scouts)
- The successive steps that are automatically performed by Brainstorm:
- Import all the surfaces (head, skull, left/right, white/pial)
- Read the anatomical label information and SVReg atlases from pial and white matter surfaces
- Downsample each hemisphere to the number specified in the options (by default 7500, half of the total default number 15000)
- Merge left and right hemispheres for the two cortical surface types: white matter and cortex envelope
- Downsample the head and skull surfaces
- Delete all the unnecessary surfaces
The files you can see in the database explorer at the end:
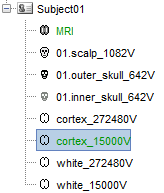
MRI: The T1 MRI of the subject
01.scalp_1082V: The head surface (downsampled to 1082 vertices)
01.outer_skull_642: The skull surface (downsampled to 642 vertices)
01.inner_skull_642: The inner skull surface (downsampled to 642 vertices)
cortex_270604V: High-resolution cortical surface that was generated by BrainSuite.
cortex_15000V: Low-resolution cortical surface, downsampled using the reducepatch function from Matlab (it keeps a meaningful subset of vertices from the original surface). This one appears in green, meaning that it is going to be used as the default by the processes that require a cortex surface.)
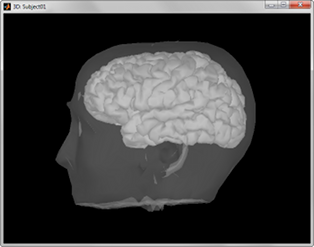
A figure is automatically shown at the end of the process, to check visually that the low-resolution cortex and head surfaces were properly imported. If it doesn't look like the following picture, do not go any further in your source analysis, fix the anatomy first.
Handling Errors
How to check the quality of the result
It's hard to estimate what would be a good cortical reconstruction. What you are trying to spot at this level is mostly the obvious errors, like when the early stages of the brain extraction didn't perform well, just with a visual inspection. Play with the Smooth slider in the Surface tab. If it looks like a brain (two separate hemispheres) in both smooth and original views, it is probably ok.
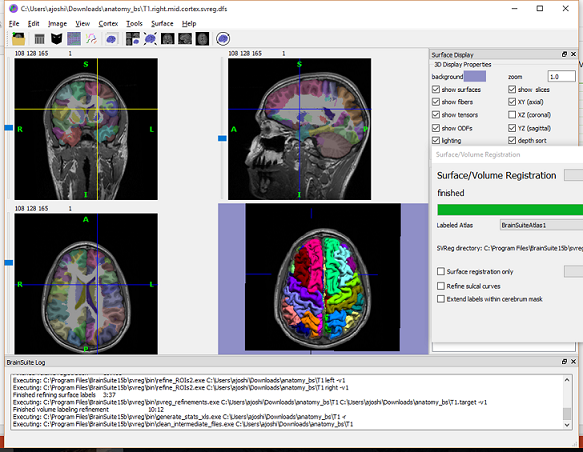
Display the cortex surface on top of the MRI slices, to make sure they are well aligned, that the surface follows the folds well, and that the left and right were not flipped: right-click on the low-resolution cortex > MRI registration > Check MRI/surface registration...
The cortex looks bad
It is critical to get a good cortex surface for source estimation. If the final cortex surface looks bad, it means that something didn't work well somewhere along the BrainSuite cortical surface extraction sequence. This tutorial is not meant to guide you though troubleshooting BrainSuite results, but most errors in surface generation and segmentation occur at the first step, skull-stripping. If you are ending up with surfaces that include non-brain regions, or are missing large chunks of cortex, this step is where the error occurred.
To fix skull-stripping issues, re-open the original T1 scan in BrainSuite. Open the Cortical Surface Extraction Sequence and check the box next to "Register and label brain" as you did above, but this time uncheck "Skull stripping". Next click on Cortex > Skull Stripping (BSE) to open the skull stripping dialog box, as this will let you step back and forth until you get a result that works. Common parameter settings that work are:
- Diffusion Constant: between 20 and 30 (inclusive)
- Edge Constant: 0.6, 0.75, 0.8
- Erosion Size: 1, 2
In general, increasing the edge constant will exclude more areas of the brain (decreasing will include more areas), while increasing the diffusion constant will include more areas. Set the parameters as you want, then click "Step >" until it says "finished skullstripping" above the progress bar. If the results look good, then click "Run All" on the Cortical Surface Extraction Sequence dialog box to run the rest of the segmentation process; if the results do not look good, click "< Back" on the Skull Stripping Tool dialog box until the progress bar returns to 0% and try a different set of parameters. More information can be found in BrainSuite's documentation.
If after following these instructions you still don't manage to get good surfaces, you can try to run the automatic MRI segmentation from BrainVISA or FreeSurfer.
The head surface looks bad
It is not mandatory to have a perfect head surface to use any of the Brainstorm features: you don't necessarily have to recognize the face (for the anonymity of the figures, it can be even better if you don't).
The head surface is important mostly for the alignment of the MEG sensors and the MRI. If you digitized the head shape with a Polhemus device, you can automatically align the head surface (hence the MRI) with the MEG sensors (in the same referential as the Polhemus points). The quality of this automatic registrations depends on the quality of both surfaces: the Polhemus head shape (green points) and the head surface from the MRI (grey surface). If you placed lots of points on the nose but your head surface doesn't have a nose, these points are not going to help. Except for that, a nice head shape is mainly useful for producing nicer figures.
If the default head surface looks bad, you can try generating another one: right-click on the subject folder > Generate head surface. The options are:
Number of vertices: Number of points that are kept from the initial isosurface computed from the MRI. Increasing this number may increase the quality of the final surface.
Erode factor: Number of pixels to erode after the first binary threshold of the MRI. Increasing this number removes small components that are connected to the head.
Fill holes factor: Number of dimensions in which the holes should be identified and closed. Increasing this number removes more of the cavities of the head surface (0=no correction, 1=removes holes inside the surface, 3=closes all the features that make the surface non-convex)
You can also try to import manually the original BrainSuite head surface. Right click on the subject > Import surface > 01.scalp.dfs. If the surface has holes, you can right-click on the head surface > Fill holes. Note that BrainSuite's head and skull files have too many vertices for analysis, so you may have to decrease the surface resolution by right-clicking the head file > Less vertices.... The automated folder importation decreases the number of vertices to 1082.
Cortical Parcellations
Checking the box next to "Register and label brain" when running BrainSuite's Cortical Surface Extraction sequence causes BrainSuite to run a companion program called SVReg that implements an automatic parcellation of the cortical surface into anatomical regions. More information on this program can be found here:http://brainsuite.bmap.ucla.edu/processing/svreg/
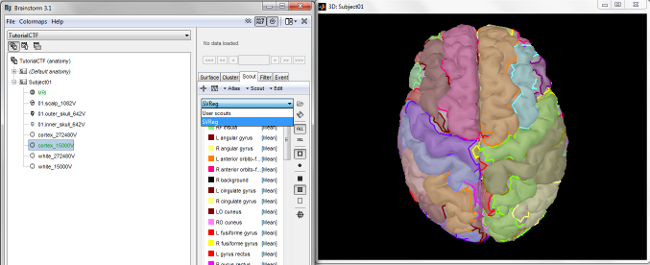
SVReg's labels are imported into BrainStorm as scouts (cortical regions of interest), and saved directly in the surface files. To check where they are saved: right-click on the low-resolution cortex file > File > View file contents. You can see that there are two "Atlas" structures available, one that has Name='User scouts' and the other with Name='SVReg'.
To access them from the interface: 1. Double-click on the low-resolution cortex (works with the original, high-resolution cortex too, it's just longer to display) 2. Go to the Scout tab, and click on the drop-down list to select "SVReg"
Troubleshooting: You do not see the atlas SVReg
If you get some error messages in the Matlab command window during the segmention ("Could not find XML label description file brainsuite_labeldescription.xml"), you are using a version of BrainSuite that does not save the file brainsuite_labeldescription.xml in the segmentation folder.
Workaround: Copy manually the file brainsuite_labeldescription.xml to the segmentation folder.
Template registration for group analysis
BrainSuite provides an accurate registration method to the Brainstorm anatomy templates (Colin27, ICBM152). The registration procedure works in the same way as the FreeSurfer-based registration illustrated in this tutorial: Group studies: Subject coregistration.
To use this registration, you need to use a template anatomy processed with BrainSuite.
Right-click on Default anatomy > Use template > Colin27_BrainSuite_2016 or ICBM152_BrainSuite_2016.