|
Size: 4358
Comment:
|
Size: 9089
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 7: | Line 7: |
== Cite FastSurfer == If you use FastSurfer for MRI segmentation, please cite the following article in your publications: Henschel L, Conjeti S, Estrada S, Diers K, Fisch B, Reuter M<<BR>>[[https://www.sciencedirect.com/science/article/pii/S1053811920304985?via=ihub|FastSurfer - A fast and accurate deep learning based neuroimaging pipeline]]<<BR>>'''NeuroImage''', Oct 2020 |
|
| Line 97: | Line 102: |
| Add the following lines at the end of the file '''$HOME/.bashrc''': {{{ # Add FastSurfer to PATH export PATH="$HOME/FastSurfer:$PATH" }}} |
|
| Line 101: | Line 112: |
| --sid PD --sd /home/ftadel/segmentation/fastsurfer_db --parallel --threads 4 | --sid PD --sd /home/ftadel/segmentation/fastsurfer_db --batch 4 --parallel --threads 4 |
| Line 104: | Line 115: |
== Run FastSurfer from Brainstorm == * Switch to the anatomy side of the database explorer. * Create a new subject, set the default anatomy option to "No, use individual anatomy". * Import the T1 MRI for this subject. * Set the [[https://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy#Fiducial_points|fiducial points]] manually (NAS/LPA/RPA) or [[https://neuroimage.usc.edu/brainstorm/Tutorials/ImportAnatomy#MNI_normalization|compute the MNI normalization]]. * Right-click on the MRI > '''MRI segmentation > FastSurfer'''. Three options can be selected interactively: the extra [[https://github.com/Deep-MI/FastSurfer#usage|command-line parameters]], the output folder (default: $HOME/.brainstorm/tmp/fastsurfer), and the number of final vertices in the final cortex surface.<<BR>><<BR>> {{attachment:fast_menu.gif}} * Alternatively, use the process: '''Import anatomy > Segment MRI with FastSurfer'''.<<BR>><<BR>> {{attachment:fast_process.gif}} * If you run the segmentation process from Brainstorm, the import will be done automatically. Otherwise, the output of the FastSurfer segmentation is compatible with FreeSurfer and can be imported with the menu Import anatomy folder, as illustrated in the [[https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer#Import_the_results_in_Brainstorm|FreeSurfer tutorial]]. * A figure is automatically shown at the end of the process, to check visually that the low-resolution cortex and head surfaces were properly generated and imported. If it doesn't look like the following picture, do not go any further in your source analysis, [[https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer#Handling_errors|fix the anatomy]] first. <<BR>><<BR>>{{attachment:fast_output.gif}} == Cortical parcellations == One of the typical [[https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer#Cortical_parcellations|FreeSurfer cortex parcellations]] is available at the end of the process: * '''Desikan-Killiany''' atlas (?h.aparc.mapped.annot): [[http://ftp.nmr.mgh.harvard.edu/fswiki/CorticalParcellation|more information]]<<BR>><<BR>>{{attachment:fast_dk.gif}} == Volume parcellations == Three of the typical [[https://neuroimage.usc.edu/brainstorm/Tutorials/LabelFreeSurfer#Volume_parcellations|FreeSurfer volume parcellation]] are available at the end of the process: * '''ASEG''': aseg.mgz * '''APARC''': aparc.mapped+aseg.mgz * '''Desikan-Killiany''': aparc.DKTatlas+aseg.deep.mgz == Subcortical structures: aseg atlas == The file aseg.mgz contains a volume atlas of 40 subcortical regions. Brainstorm reads these volume labels and tesselates some of these regions, groups all the meshes in a large surface file where the regions are identified in an atlas called "Structures". It identifies: 8 bialateral structures (accumbens, amygdala, caudate, hippocampus, pallidum, putamen, thalamus, cerebellum) and 1 central structure (brainstem). You can easily extract one structure (for example the brainstem or the cerebellum) by selecting the corresponding entries in the scouts list and selecting the menu Scout > Edit surface > Keep only selected scouts. It creates a new surface with only the selected regions. If you want to remove one or several structures, use the menu "Remove selected scouts" instead. [[https://neuroimage.usc.edu/brainstorm/Tutorials/DeepAtlas#Select_deep_structures|More information]]. Read more about the FreeSurfer subcortical atlas on the software wiki: http://ftp.nmr.mgh.harvard.edu/fswiki/SubcorticalSegmentation == Registered spheres == The registered spheres are saved in each surface file in the field Reg.Sphere.Vertices. There is nothing that can be done with this information at this point, but it will become helpful when projecting the source results from the individual brains to the default anatomy of the protocol, for a group analysis of the results: [[https://neuroimage.usc.edu/brainstorm/Tutorials/CoregisterSubjects|Subject coregistration]]. Read more about the FreeSurfer registration process on the software wiki: https://surfer.nmr.mgh.harvard.edu/fswiki/SurfaceRegAndTemplates |
MRI segmentation with FastSurfer
Authors: Francois Tadel
The open-source software FastSurfer can be used to extract the cortical envelope from a T1 MRI and register it to an atlas. The process is fully automatic, the results are compatible with FreeSurfer. If you are using FastSurfer, please cite the appropriate references. Note that this software is available only for Linux and MacOS.
Contents
Cite FastSurfer
If you use FastSurfer for MRI segmentation, please cite the following article in your publications:
Henschel L, Conjeti S, Estrada S, Diers K, Fisch B, Reuter M
FastSurfer - A fast and accurate deep learning based neuroimaging pipeline
NeuroImage, Oct 2020
Install FastSurfer
The most complicated task will be to install FastSurfer. Unfortunately, Brainstorm cannot do that automatically yet. If you are using Ubuntu 20, you may try the procedure below. Otherwise, refer to the intructions in the FastSurfer GitHub repository.
Install pyenv: Python 3.6
From a terminal, install the required packages:
$ sudo apt-get install -y make build-essential libssl-dev zlib1g-dev \ libbz2-dev libreadline-dev libsqlite3-dev wget curl llvm libncurses5-dev \ libncursesw5-dev xz-utils tk-dev libffi-dev liblzma-dev python-openssl
Install pyenv:
$ curl https://pyenv.run | bash
Add the following lines at the end of the file $HOME/.bashrc:
# Load pyenv-virtualenv automatically export PATH="$HOME/.pyenv/bin:$HOME/.pyenv/shims:$PATH" eval "$(pyenv init -)"eval "$(pyenv virtualenv-init -)"
Start a new terminal, install Python 3.6 and create a new virtual environment for FastSurfer:
$ pyenv install 3.6.15 $ pyenv global 3.6.15 $ pyenv virtualenv fastsurfer
For complete and updated installation instruction, see the pyenv repository.
Install FreeSurfer 6.0.1
Note that FastSurfer works only with FreeSurfer 6. Newer versions of FreeSurfer are not supported at the moment. If you try using FreeSurfer 7, you would get an error message when running recon-surf.
Install the required packages:
$ apt-get -y install bc binutils libgomp1 perl psmisc sudo tar tcsh unzip \
uuid-dev vim-common libjpeg62-devDownload and install FreeSurfer 6.0.1:
$ cd ~/ $ wget https://surfer.nmr.mgh.harvard.edu/pub/dist/freesurfer/6.0.1/freesurfer-Linux-centos6_x86_64-stable-pub-v6.0.1.tar.gz $ tar xzvf freesurfer-Linux-centos6_x86_64-stable-pub-v6.0.1.tar.gz $ sudo mkdir /usr/local/freesurfer $ sudo mv freesurfer /usr/local/freesurfer/6.0.1
Obtain a license.txt file from this page, and copy it to the installation folder:
$ cp license.txt /usr/local/freesurfer/6.0.1/
Add the following lines at the end of the file $HOME/.bashrc:
# FreeSurfer export FREESURFER_HOME=/usr/local/freesurfer/6.0.1/ source $FREESURFER_HOME/SetUpFreeSurfer.sh
Close the terminal and open a new one, you should see the following message:
-------- freesurfer-Linux-centos6_x86_64-stable-pub-v6.0.1-f53a55a -------- Setting up environment for FreeSurfer/FS-FAST (and FSL) FREESURFER_HOME /usr/local/freesurfer/6.0.1 FSFAST_HOME /usr/local/freesurfer/6.0.1/fsfast FSF_OUTPUT_FORMAT nii.gz SUBJECTS_DIR /usr/local/freesurfer/6.0.1/subjects MNI_DIR /usr/local/freesurfer/6.0.1/mni
For complete and updated installation instructions, see the FreeSurfer wiki.
Install FastSurfer
Clone the FastSurfer GitHub repository:
$ cd ~/ $ git clone https://github.com/Deep-MI/FastSurfer.git
Install the required packages:
$ sudo apt install libblas-dev liblapack-dev libatlas-base-dev gfortran $ cd FastSurfer $ pip install -r requirements.txt
Add the following lines at the end of the file $HOME/.bashrc:
# Add FastSurfer to PATH export PATH="$HOME/FastSurfer:$PATH"
Execute FastSurfer manually:
$ ./run_fastsurfer.sh --t1 /home/ftadel/segmentation/freesurfer_db/PD/mri/orig/001.mgz \ --sid PD --sd /home/ftadel/segmentation/fastsurfer_db --batch 4 --parallel --threads 4
This default configuration would run FastSurfer on the CPU. If you have access to GPU resources and have the CUDA libraries installed on your system, you could make FastSurfer run much faster. See the documentation on the FastSurfer GitHub repository.
Run FastSurfer from Brainstorm
- Switch to the anatomy side of the database explorer.
- Create a new subject, set the default anatomy option to "No, use individual anatomy".
- Import the T1 MRI for this subject.
Set the fiducial points manually (NAS/LPA/RPA) or compute the MNI normalization.
Right-click on the MRI > MRI segmentation > FastSurfer. Three options can be selected interactively: the extra command-line parameters, the output folder (default: $HOME/.brainstorm/tmp/fastsurfer), and the number of final vertices in the final cortex surface.
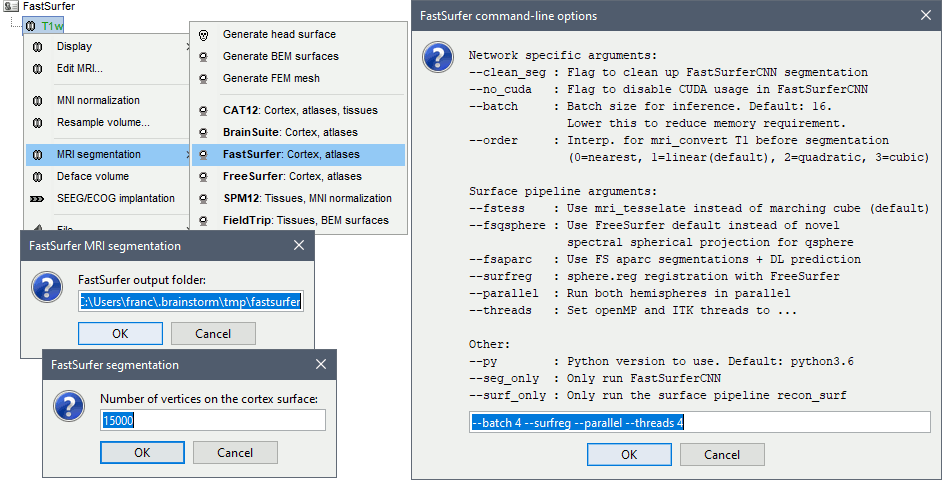
Alternatively, use the process: Import anatomy > Segment MRI with FastSurfer.
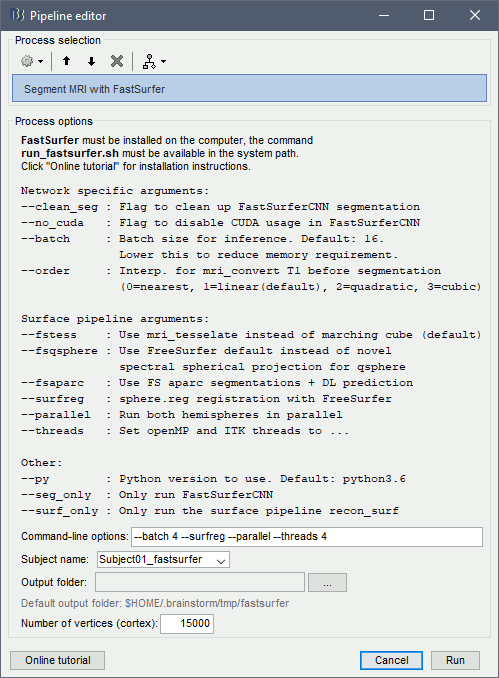
If you run the segmentation process from Brainstorm, the import will be done automatically. Otherwise, the output of the FastSurfer segmentation is compatible with FreeSurfer and can be imported with the menu Import anatomy folder, as illustrated in the FreeSurfer tutorial.
A figure is automatically shown at the end of the process, to check visually that the low-resolution cortex and head surfaces were properly generated and imported. If it doesn't look like the following picture, do not go any further in your source analysis, fix the anatomy first.
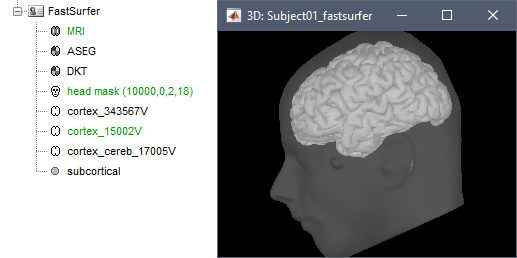
Cortical parcellations
One of the typical FreeSurfer cortex parcellations is available at the end of the process:
Desikan-Killiany atlas (?h.aparc.mapped.annot): more information
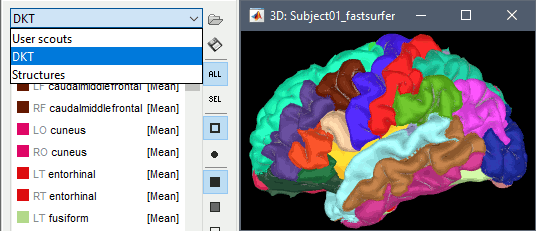
Volume parcellations
Three of the typical FreeSurfer volume parcellation are available at the end of the process:
ASEG: aseg.mgz
APARC: aparc.mapped+aseg.mgz
Desikan-Killiany: aparc.DKTatlas+aseg.deep.mgz
Subcortical structures: aseg atlas
The file aseg.mgz contains a volume atlas of 40 subcortical regions. Brainstorm reads these volume labels and tesselates some of these regions, groups all the meshes in a large surface file where the regions are identified in an atlas called "Structures". It identifies: 8 bialateral structures (accumbens, amygdala, caudate, hippocampus, pallidum, putamen, thalamus, cerebellum) and 1 central structure (brainstem).
You can easily extract one structure (for example the brainstem or the cerebellum) by selecting the corresponding entries in the scouts list and selecting the menu Scout > Edit surface > Keep only selected scouts. It creates a new surface with only the selected regions. If you want to remove one or several structures, use the menu "Remove selected scouts" instead. More information.
Read more about the FreeSurfer subcortical atlas on the software wiki:
Registered spheres
The registered spheres are saved in each surface file in the field Reg.Sphere.Vertices. There is nothing that can be done with this information at this point, but it will become helpful when projecting the source results from the individual brains to the default anatomy of the protocol, for a group analysis of the results: Subject coregistration.
Read more about the FreeSurfer registration process on the software wiki:
