SEEG Time-Frequency Analysis for Epileptogenic Zone Localization (under construction)
Authors: xxx.
Detection, localization and labelling of SEEG depth electrodes is a vital step in studying brain activity.
Note that the operations used here are not detailed, the goal of this tutorial is not to introduce Brainstorm to new users. For in-depth explanations of the interface and theoretical foundations, please refer to the introduction tutorials.
NOT FOR CLINICAL USE:
The performance characteristics of the methods and software implementation presented in this tutorial have not been certified as medical devices and should be used for research purposes only.
Dataset description
License
This tutorial dataset (SEEG and MRI data) remains property of the Cleveland Clinic, Ohio, USA. Its use and transfer outside the Brainstorm tutorials, e.g. for research purposes, is prohibited without written consent. For questions, please contact Kenneth Taylor, PhD ( taylork2@ccf.org ).
SEEG recordings
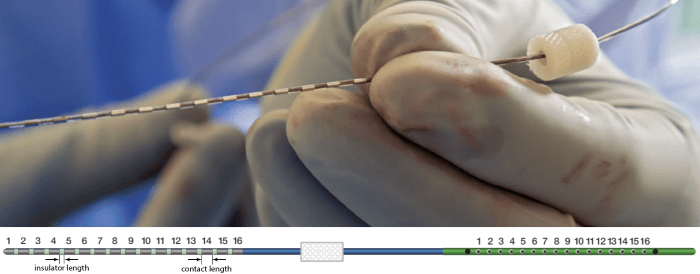
The depth electrodes used in this example dataset are PMT SEEG Depth Electrodes, with the following specifications:
- Diameter: 0.8 mm
- Contact length: 2 mm
- Insulator length: 1.5 mm
- Distance between the center of two contacts: 3.5 mm
- Between 8 and 16 contacts on each electrode
Files
tutorial_seeg_implantation/
raw/: All the raw MRI and CT scans.
seeg_straight/: Bent electrodes not available for this subject
preMRI.nii: MRI before the implantation of the SEEG.
postCT.nii: CT after the implantation of the SEEG.
seeg_curved/: Bent electrodes available for this subject
preMRI_curved.nii: MRI before the implantation of the SEEG.
postCT_curved.nii: CT after the implantation of the SEEG.
precomputed/TutorialSeegImplantation_precomputed.zip: The precomputed Brainstorm protocol for this tutorial (Do not unzip this file).
- All the anatomical images have been de-identified with defacing from Brainstorm.
Download and installation
Requirements: You have already followed all the introduction tutorials and you have a working copy of Brainstorm installed on your computer.
BrainSuite (optional): Make sure you have the latest version of BrainSuite installed. Follow steps as per the BrainSuite for Brainstorm tutorial. This is required to perform skull stripping when importing CT below in order to remove extracranial regions from the CT.
SPM: If you are running Brainstorm from the MATLAB environment, you need to have the SPM12 toolbox installed on your computer, as a Brainstorm plugin or a custom installation.
With the stand-alone compiled version of Brainstorm: all the needed SPM scripts have been compiled and included in the executable. This is required for two purposes:- Coregistration between pre-implantation MRI and post-implantation CT volumes.
Use mask from SPM based tissue segmentation to remove extracranial regions from the CT.
ct2mrireg: To have an alternate way of doing the coregistration between pre-implantation MRI and post-implantation CT volumes, you need to have the ct2mrireg plugin installed on your computer, as a Brainstorm plugin. It can be found under the menu Plugins > Anatomy > ct2mrireg. If using Brainstorm with MATLAB, please install the Image Processing Toolbox.
Download the dataset:
Go to the Download page of this website, and download the file: tutorial_seeg_implantation.zip.
- Unzip it in a folder that is not in any of the Brainstorm folders (program folder or database folder).
Brainstorm:
- Start Brainstorm
Select the menu File > Create new protocol. Name it "TutorialSeegImplantation" and select the options:
"No, use individual anatomy",
"No, use one channel file per acquisition run".