|
Size: 361
Comment:
|
Size: 45257
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Tutorial 17: Time-frequency = ''Authors: Francois Tadel, ''''''Dimitrios Pantazis, ''Elizabeth Bock, Sylvain Baillet'' |
= Tutorial 24: Time-frequency = '''[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE] ''' ''Authors: Francois Tadel, Dimitrios Pantazis, Elizabeth Bock, Sylvain Baillet'' This tutorial introduces how to compute time-frequency decomposition of MEG/EEG recordings and cortical currents using complex Morlet wavelets and Hilbert transforms. |
| Line 6: | Line 10: |
| <<EmbedContent("http://neuroimage.usc.edu/bst/get_prevnext.php?prev=Tutorials/Scouts&next=Tutorials/Statistics")>> | == Introduction == Some of the MEG/EEG signal properties are difficult to access in time domain (graphs time/amplitude). A lot of the information of interest is carried by oscillations at certain frequencies, but the amplitude of these oscillations is sometimes a lot lower than the amplitude of the slower components of the signal, making them difficult to observe. The averaging in time domain may also lead to a cancellation of these oscillations when they are not strictly locked in phase across trials. Averaging trials in time-frequency domain allows to extract the power of the oscillation regardless of the phase shifts. For a better understanding of this topic, we recommend the lecture of the following article: Bertrand O, Tallon-Baudry C (2000), [[http://www.iec-lnc.ens.fr/IMG/Files/Visual/Publications/2000_Review_IntJPsychol.pdf|Oscillatory gamma activity in humans: a possible role for object representation]] In Brainstorm we offer two approaches for computing time-frequency decomposition (TF): the first one is based on the convolution of the signal with series of complex Morlet wavelets, the second filters the signal in different frequency bands and extracts the envelope of the filtered signals using the Hilbert transform. {{attachment:slide.gif}} == Morlet wavelets == Complex Morlet wavelets are very popular in EEG/MEG data analysis for time-frequency decomposition. They have the shape of a sinusoid, weighted by a Gaussian kernel, and they can therefore capture local oscillatory components in the time series. An example of this wavelet is shown below, where the blue and red curves represent the real and imaginary part, respectively. Contrary to the standard short-time Fourier transform, wavelets have variable resolution in time and frequency. For low frequencies, the frequency resolution is high but the time resolution is low. For high frenquies, it's the opposite. When designing the wavelet, we basically decide a trade-off between temporal and spectral resolution. . {{attachment:tf_resolution.gif||height="144",width="336"}} To design the wavelet, we first need to choose a ''central frequency'', ie. the frequency where we will define the mother wavelet. All other wavelets will be scaled and shifted versions of the mother wavelet. Unless interested in designing the wavelet at a particular frequency band, the default 1Hz should be fine. Then, the desirable'' time resolution'' for the central frequency should be defined. For example, we may wish to have a temporal resolution of 3 seconds at frequency 1 Hz (default parameters). These two parameters, uniquely define the temporal and spectral resolution of the wavelet for all other frequencies, as shown in the plots below. Resolution is given in units of Full Width Half Maximum of the Gaussian kernel, both in time and frequency. . {{attachment:waveletOptions.gif||width="650px"}} == Edge effects == Users should pay attention to edge effects when applying wavelet analysis. Wavelet coefficients are computed by convolving the wavelet kernel with the time series. Similarly to any convolution of signals, there is zero padding at the edges of the time series and therefore the wavelet coefficients are weaker at the beginning and end of the time series. From the figure above, which designs the Morlet wavelet, we can see that the default wavelet (central frequency '''fc'''=1Hz, '''FWHM_tc'''=3sec) has temporal resolution 0.6sec at 5Hz and 0.3sec at 10Hz. In such case, the edge effects are roughly half these times: 300ms in 5Hz and 150ms in 10Hz. More precisely, if '''f''' is your frequency of interest, you can expect the edge effects to span over FWHM_t seconds: '''FWHM_t = FWHM_tc * fc / f / 2'''. Examples of such transients are given in the figures below. {{attachment:edgeEffect5Hz.gif||height="244",width="299"}} {{attachment:edgeEffect10Hz.gif||height="245",width="302"}} == Simulation == We will illustrate the time-frequency decomposition process with a simulated signal. * The following code generates a sum of three sinusoids (2Hz, 20Hz, 50Hz) with random white noise. The 50Hz and noise are present everywhere, the 2Hz and 20Hz start only after two seconds.<<BR>> {{{ f1 = 2; f2 = 20; f3 = 50; i =2000:6000; Data(1,i) = sin(f1*2*pi*t(i)) + 0.4 * cos(f2*2*pi*t(i)); Data = Data + 0.2 * sin(f3*2*pi*t) + 0.4 * rand(1,6000); }}} * Empty the Process1 list (right-click > Clear list) then click on [Run]. * Run process: '''Simulate > Simulate generic signals'''. <<BR>>Ntime='''6000''', Sampling frequency='''1000Hz''' (signal duration = 6000/1000 = 6 seconds).<<BR>>Copy-paste the few lines of code above to generate the sum of three sinusoids.<<BR>><<BR>> {{attachment:simulate_process.gif||height="412",width="489"}} * Double-click on the new file to look at the simulated signal.<<BR>><<BR>> {{attachment:simulate_display.gif||height="159",width="667"}} * In Process1, select the simulated signal. * Run process: '''Frequency > Time-frequency (Morlet wavelets)'''. <<BR>>Select the option '''1/f compensation''': The normalization will be discussed later.<<BR>><<BR>> {{attachment:example_process.gif||height="271",width="557"}} * Click on the button ['''Edit'''] to see all the process options.<<BR>>Time definition: '''Same as input''', Frequency definition: '''Linear 1:1:60''', Compute measure: '''Power'''. <<BR>><<BR>> {{attachment:example_options.gif||height="542",width="403"}} == Process options == '''Comment''': String that will be displayed in the database explorer to represent the output file. '''Time definition''' * '''Same as input file''': The output file has the same time definition as the input file. <<BR>>In this example, it means: 6000 samples between 0 and 6s. * '''Group in time bands''': This option adds a step of computation. First it computes the TF decomposition for all the input file, then averages the power by time band. To define a time band: * Enter your own time bands in the text area, one line per time band, with the following format: '''"name / time definition / function" ''' * Click on the button [Generate] to automatically create a list of time bands with the same length. You will be asked the maximal length of each time band. * The function is the measure we take to combine the values for all the individual frequencies into one for the frequency band. Possible values are: '''mean, max, std, median'''. '''Frequency definition''': Frequencies for which the power will be estimated at each time instant. * '''Linear''': You can specify the frequencies with the Matlab syntax start:step:stop. <<BR>>The default is "1:1:60", which produces 60 values [1, 2, 3, 4, ..., 59, 60]. * '''Log''': With the option start:N:stop, produces a list of N frequencies logarithmically scaled between "start" and "stop". For example "1:40:80" is converted to [1, 1.5, 2.1, 2.7, ..., 61.5, 65.8, 75, 80] * '''Group in frequency bands''': As for the time definition, this option leads to a two-step process. First it computes the TF decomposition for several values in the frequency band, then it averages the power of TF coefficients per frequency band. To define a frequency band: * One line per frequency band, with the format '''"name / frequency definition / function" ''' * The frequency definition is a Matlab expression evaluated with an eval() call. If the frequency definition contains only two values, Brainstorm adds two extra values in the middle so that the final averaged value is a bit more robust. Example of valid expressions:<<BR>>"'''2,4'''": Evaluates to [2,4], and then expands to the frequency vector [2, 2.66, 3.33, 4]<<BR>>"'''2:0.5:4'''": Evaluates to [2 2.5 3 3.5 4]<<BR>>"'''2, 2.5, 3, 3.5, 4'''": Evaluates to [2 2.5 3 3.5 4] * The function is the measure we take to combine the values for all the individual frequencies into one for the frequency band. Possible values are: '''mean, max, std, median'''. '''Morlet wavelet options''' * '''Central frequency''': Frequency where the mother wavelet is designed. All other wavelets will be shifted and scaled versions of the mother wavelet * '''Time resolution (FWHM)''': Temporal resolution of the wavelet at the central frequency (in units of Full Width Half Maximum). Click [Display] to see the resolution of the wavelet for other frequencies. '''Compute the following measure''': * The convolution of the signal with complex Morlet wavelets returns the complex coefficients for each frequency/time/sensor. Typically, what we display is the power of the coefficients (square of the amplitude: abs(TF).^2). You can choose if you want to apply this transformation or not. * '''Power''': Computes the "power" transformation immediately after the TF decomposition. This discards the phase information, but produces files that are twice smaller and a lot easier to process. * '''None''': Save the TF coefficients as they are computed. This can be useful if you plan to use these decompositions for other purposes that require the phase. * Some combinations of options may disable this choice. If you select frequency bands, the program will have to compute the power before averaging the values, therefore "none" is not an option. == Display: Time-frequency map == * Right on the new time-frequency file > '''Time-freq: One matrix''' (same as double-clicking). <<BR>>This menu displays the time-frequency decomposition of the first (and only) signal. The Brainstorm window shows two new elements: the tab "Display" and the frequency slider.<<BR>><<BR>> {{attachment:example_file.gif||height="210",width="660"}} * We can easily identify the three horizontal bars as the three sinusoids in the simulated signals, and the trade-off between accuracy in time and accuracy in frequency. Click on the figure to move the time-frequency cursor and explore the two axes of the plane. * '''2Hz''': High frequency resolution but poor time resolution (supposed to start sharply at 2s) * '''20Hz''': Better time resolution but poorer frequency resolution (17-24Hz) * '''50Hz''': Continuous over the 6s - Frequency resolution gets even worse (40-60Hz). It looks discontinuous because this oscillation has the same amplitude as the white noise we added in the signal (weight 0.4, relatively to the 2Hz oscillation). * '''Current frequency''': Slider that shows the current frequency selected in all the figures. <<BR>>Just like the time, the frequency selection is centralized and managed by one control only for all the figures. As a consequence, it is impossible to display TF files with different frequency definitions at the same time. This can be perceived as an annoying limitation, but it allows all the simultaneous displays to be consistent at anytime and makes the interface more intuitive to manipulate, with lower risks of mistakes in the interpretation of the different figures. * '''List of signals''': This drop-down list shows the signal currently displayed in the selected figure. In this case, there is only one channel of data called "s1". It will be more useful later. * '''Hide edge effects''': When this option is selected, the time-frequency coefficients that could not be properly estimated because of a lack of time samples are hidden. It allows you to see only the information that is really reliable. The lower the frequency, the longer the edge effects.<<BR>><<BR>> {{attachment:example_hide.gif||height="207",width="555"}} * '''Smooth display''': Re-interpolates the time-frequency maps on a finer grid to produce nicer plots. * '''Measure''': Type of measure that is currently represented in the selected figure. The entries that are enabled depend on the type of data that is saved in the file. In this case, we saved directly the power of the wavelet coefficients in the file, we discarded the angle/phase information, so the "phase" option is disabled. The other options are: '''Magnitude''' = sqrt(power), '''Log''' = 10*log10(power) * '''Colormap''': As explained in the previous tutorials, you can change the colormap by clicking+moving on the colorbar on the right of the figure. Double-click on the colorbar to restore the defaults. == Display: Mouse and keyboard shortcuts == ==== Mouse shortcuts ==== * '''Left-click''': Selection of current time and frequency. * '''Left-click + move''': Select a time/frequency range. The legends of the X/Y axis show the selection. <<BR>><<BR>> {{attachment:example_select.gif||height="159",width="362"}} * '''Mouse wheel''': Zoom in time, centered on the current time cursor. * '''Control + mouse wheel''': Zoom in frequencies, centered on the current frequency cursor. * '''Right-click + move''', or Control + left-click + move: Move in the zoomed image. * '''Double-click''': Restore initial view. ==== Keyboard shortcuts: ==== * '''Left/right arrows''': Change the current time. * '''Page-up/page-down''': Change the current time, 10 time samples at a time. * '''Up/down arrows''': Change the the sensor displayed in this figure. * '''Control + up/down arrows''': Change the current frequency. * '''Enter''': View the original time series for this sensor. * '''Control + R''': View the original MEG recordings. * '''Control + T''': View the time-frequency 2D topography. * '''Control + I''': Save as image. * '''Control + D''': Dock figure in the Matlab environment. ==== Figure popup menu ==== . {{attachment:example_popup.gif||height="191",width="480"}} * '''Set selection manually''': Does the same thing as drawing a time/freq selection square on a figure, but by typing the values for time and frequency manually. * '''Export to database''': Save the selection for the displayed sensor in a new file in the database. * '''Export to file''': Same as "Export to database", but the saved file is not registered in the database. * '''Export to Matlab''': Same as "Export to database", but the output structure is sent to a variable in the Matlab base workspace instead of being saved to a file. == Display: Power spectrum and time series == These two menus generate similar figures: * '''Power spectrum''': For the current time, shows the power for all the frequencies. * '''Time series''': For the current frequency, shows the power for all the time samples. * Example: Power spectrum density at '''0.5s''' and power time series at '''2Hz'''. <<BR>>We see the oscillation at the 50Hz in the PSD plot, and the oscillation at 2Hz in the TS plot.<<BR>><<BR>> {{attachment:example_psd1.gif||height="148",width="660"}} * Example: Power spectrum density at '''4s''' and power time series at '''20Hz'''. <<BR>>We see all three oscillations in the PSD plot, and the oscillation at 20Hz in the TS plot.<<BR>><<BR>> {{attachment:example_psd2.gif||height="148",width="661"}} * Note that if you right-click on the file in the '''database explorer''' and then select one of these menus, it will show '''all the signals'''. If you right-click on an existing '''time-frequency figure''', it will show only the '''selected signal'''. It doesn't make any difference here because there is only one signal, but it will with the MEG recordings. == Normalized time-frequency maps == The brain is always active, the MEG/EEG recordings are never flat, some oscillations are always present in the signals. Therefore we are often more interested in the transient changes in the power at certain frequencies than at the actual power. A good way to observe these changes is to compute a '''deviation of the power with respect with a baseline'''. There is another reason for which we are usually interested in standardizing the TF values. The power of the time-frequency coefficients are always lower in the higher frequencies than in the lower frequencies, the signal carries a lot less power in the faster oscillations than in the slow brain responses. This '''1/f decrease in power''' is an observation that we already made with the power spectrum density in the [[Tutorials/ArtifactsFilter|filtering tutorial]]. If we represent the TF maps with a linear color scale, we will always see values close to zero in the higher frequency ranges. Normalizing each frequency separately with respect with a baseline helps obtaining more readable TF maps. ==== No normalization ==== The values we were looking at were already normalized (option: "1/f compensation" in the process options), but not with respect to a baseline. We will now compute the non-normalized power obtained with the Morlet wavelets and try the various options available for normalizing them. * In Process1, keep the simulated signal selected. * Run process: '''Frequency > Time-frequency (Morlet wavelets)''', "Spectral flattening: '''None'''"<<BR>><<BR>> {{attachment:example_process_none.gif||height="223",width="458"}} * Double-click on the file. As expected, we only see the lower frequencies in this representation: the power of the 2Hz oscillation is a lot larger than the power at 20Hz or 60Hz. <<BR>><<BR>> {{attachment:example_display_none.gif||height="131",width="433"}} ==== Spectral flattening ==== * In Process1: Select this new non-normalized file "Power,1-60Hz". * Run process: '''Standardize > Spectral flattening''', Method='''1/f compensation'''. <<BR>><<BR>> {{attachment:norm_multiply.gif||height="224",width="454"}} * This produces exactly the same results as previously (option "1/f compensation" in the time-frequency process). It '''multiplies the power''' at each frequency bin '''with the frequency''' value (eg. multiplies by 20 the power at 20Hz), in order to correct for the 1/f shape we observe in the power spectrum. This works well for the lower part of the spectrum and up to 60-80Hz, but past this range it tends to '''overcompensate the higher frequencies'''.<<BR>>Note that it does not do any form of baseline correction: the 50Hz oscillation is visible everywhere. ==== Baseline normalization ==== * The second way to proceed is to normalize the power with respect to its average level during a reference time period. We can consider the oscillations at 2Hz and 20Hz as our events of interest, and the 50Hz as noise we want to get rid of. The segment from '''0 to 2 seconds''' does not contain any of the signals of interest, therefore we can consider it as a '''baseline'''. * However, we will not be able to use the full segment [0,2]s because of the edge effects we described at the beginning of this tutorial. The time-frequency map at '''2Hz''' with the display option "Hide edge effects" (left figure below) shows that the power could not be estimated correctly before 0.75s, therefore we shouldn't use it as a baseline. The power time series at 2Hz (right) shows that the power related with the 2Hz oscillation starts to increase significantly after 1.25s, therefore it's not really a "baseline" anymore. This leaves only the segment '''[0.75,1.25]s''' available. <<BR>><<BR>> {{attachment:norm_baseline_2hz.gif||height="195",width="551"}} * At '''20Hz''', the expected transient effects are only 75ms long, therefore we could use a much longer baseline if we were not interested by the lower frequencies: '''[0.075, 1.925]s'''. <<BR>><<BR>> {{attachment:norm_baseline_20hz.gif||height="195",width="551"}} * In this case, we have a very long "resting" time segment (2s), therefore the edge effects are not a problem for picking a time window for the baseline normalization. We will use the first time window mentioned, [0.75,1.25]s as it is long enough (500ms) to estimate the baseline mean power. In real-life cases, with shorter epochs, it is sometimes difficult to find an acceptable '''trade-off between the baseline duration and the exclusion of the edge effects''', especially for the lower frequencies. * Run process: '''Standardize > Baseline normalization''', Baseline='''[0.75, 1.25]s'''.<<BR>>Method='''Event-related perturbation''': ERS/ERD stands for "event-related synchronization / desynchronization", a widely used normalization measure for time-frequency power maps. It evaluates the deviation from the mean over the baseline, in percents: '''(x-mean)/mean*100'''. <<BR>><<BR>> {{attachment:norm_ersd_process.gif||height="341",width="477"}} * Double-click on the new "ersd" file. The colormap type changed from "'''Timefreq'''" to "'''Stat2'''", which uses by default the "'''rwb'''" color set and shows relative values. Indeed, the ERS/ERD values can be '''positive or negative''': the power at a given time sample can be higher or lower than the average level during the baseline. In the simple simulation we used, there is no power decrease at any frequency after 2s, so the strong values are mostly positive. However, if you look in the file, you would see that there are many small negative values (due to the random noise we added). <<BR>><<BR>> {{attachment:norm_ersd_file.gif||height="187",width="499"}} * Note that the '''50Hz disappeared''' because it was present during the baseline, while the 2Hz and 20Hz oscillations show high positive values (between 100% and 2000% increase from baseline). * Remember to select your baseline very carefully according to the frequency range you are interested in. See below examples obtained with different baselines: '''0.075-1.925s''', '''0-2s''', '''0-6s'''. <<BR>>Change the colormap to "jet" if you prefer, and adjust the colormap contrast as needed. <<BR>><<BR>> {{attachment:norm_ersd_compare.gif||height="202",width="661"}} <<TAG(Advanced)>> == Tuning the wavelet parameters == ==== Time resolution ==== You can adjust the relative time and frequency resolution of your wavelet transformation by adjusting the parameters of the mother wavelet in the options of the process. * Increasing the option "time resolution" will produce longer wavelets at a given frequency, hence increase the frequency accuracy (lower <<HTML(Δ)>>f) and decrease the time accuracy (higher <<HTML(Δ)>>t). Expect longer edge effects. * Decrease the time resolution will produce shorter wavelets at a given frequency, hence decrease the frequency accuracy (higher <<HTML(Δ)>>f) and increase the time accuracy (higher <<HTML(Δ)>>t). Expect shorter edge effects. * You can modify one or the other parameter, what is important is the '''product of the two'''. All the following combinations fc/FWHM_t produce the same results because their product is constant: (1Hz,3s), (3Hz,1s), (6Hz,0.5s), (60Hz,0.05s) <<BR>><<BR>> {{attachment:tf_resolution_process.gif}} Examples for a constant central frequency of '''1Hz''' with various time resolutions: '''1.5s''', '''4s''', '''10s'''. . {{attachment:tf_resolution_compare.gif||height="183",width="599"}} ==== Frequency axis ==== You can also obtain very different representations of the data by changing the '''list of frequencies''' for which you estimate the power. You can also change this in the options of the process. . {{attachment:tf_freqlist_process.gif}} Examples: Log 1:20:150, Log 1:300:150, Linear 15:0.1:25 . {{attachment:tf_freqlist_compare.gif||height="183",width="598"}} <<TAG(Advanced)>> == Hilbert transform [TODO] == We can repeat the same analysis with the other approach available for exploring the simulated signal in the time-frequency plane. The process "'''Frequency > Hilbert transform'''" first filters the signals in various frequency bands with a band-pass filter, then computes the Hilbert transform of the filtered signal. The magnitude of the Hilbert transform of a narrow-band signal is a measure of the envelope of this signal, and therefore gives an indication of the activity in this frequency band. . {{attachment:slide_hilbert.gif}} ==== No normalization ==== Let's compute the same three results as before: non-normalized, 1/f compensation, baseline normalization. * In Process1, select the simulated signal. * Run process: '''Frequency > Hilbert transform''', Spetral flattening='''None''', '''Mirror signal''' before. <<BR>> The mirroring option helps with controlling the amplitude of the edge effects.<<BR>><<BR>> {{attachment:hilbert_process.gif||height="242",width="460"}} * In the advanced options panel, keep the default options: Default frequency bands and '''Magnitude'''.<<BR>><<BR>> {{attachment:hilbert_options.gif||height="400",width="388"}} * Double-click on the new file. The figure now has only 6 rows, one for each frequency band. <<BR>>Note that the values saved in the file and displayed are now '''amplitude''' and not power anymore. <<BR>><<BR>> {{attachment:hilbert_file.gif||height="197",width="606"}} * The non-normalized results are already easy to interpret: * '''delta (2-4Hz)''': Includes the 2Hz oscillation, contribution starts at 2s * '''beta (15-29Hz)''': Includes the 20Hz oscillation, contribution starts at 2s * '''gamma1(30-59Hz)''': Includes the 50Hz oscillation, contribution starts at the beginning (0s) * Right-click on the file or figure > Time series. Example for delta and beta.<<BR>><<BR>> {{attachment:hilbert_ts.gif||height="136",width="429"}} ==== Normalization ==== * In Process1, select the non-normalized Hilbert-based decomposition. * Run process: '''Standardize > Spectral flattening''', Method='''1/f compensation'''. * Run process: '''Standardize > Baseline normalization''', Baseline='''[0.75, 1.25]s''', Method='''ERS/ERD'''. * Display the two normalized files side by side. <<BR>><<BR>> {{attachment:hilbert_normalized.gif||height="155",width="609"}} ==== Method specifications ==== * Filters: '''[TODO]''' * Hilbert transformation: Using the [[http://www.mathworks.com/help/signal/ref/hilbert.html|Matlab's hilbert()]] function. * Extraction of the envelope: Magnitude of the complex Hilbert transformat, abs(hilbert(x)). ==== Magnitude vs power ==== * Be careful with the direct comparison you obtain between this approach and the previous one. By default, the '''Hilbert''' process saves everything in '''amplitude''' (magnitude of the Hilbert transform) while the '''Morlet '''wavelet process saves everything in '''power''' (square of the wavelet coefficients). * This follows the common practices in two different fields of signal processing: envelopes are typically displayed in amplitude, Fourier-transform and other spectral decompositions are displayed in power. * So even if it looks like these two processes produce similar outputs, they don't. Make sure you take into account this difference in your statistical analysis and reports. == MEG recordings: Single trials == Now let's go back to our auditory oddball paradigm and apply the concepts to MEG recordings. We will use all the trials available for one condition to estimate the average time-frequency decomposition. ==== Spectral flattening ==== * In Process1, select all the '''deviant trials''' in '''Run#01'''. * Run process: '''Frequency > Time-frequency (Morlet wavelets)''', Spectral flattening: '''None'''. <<BR>>In the advanced options, select: '''Log 1:40:150''', '''Power''', Save '''average''' time-frequency maps. <<BR>><<BR>> {{attachment:tf_trials_process.gif||height="214",width="352"}} {{attachment:tf_trials_options.gif||height="318",width="300"}} * '''Save individual TF maps''': This option stops the computation here and saves in the database one time-frequency file for each input file (40 files), with one TF map for each scout. * '''Save average TF maps''': Instead of saving the TF for each file separately, it automatically computes the average of the power of all the TF. This is a good choice if you do not plan to use independently all the TF files, because it saves a lot of time and disk space. * '''Remove evoked response from each trial before computing TF''': This option first computes the average of all the trials, then subtracts this average from each trial before computing the time-frequency decomposition. This brings the signals to a slightly more stationnary state, which may help for the evaluation of the frequency contents. ==== Baseline normalization ==== * Double-click on the new file '''Avg,Power,1-150Hz (MEG)'''. Select "'''Hide edge effects'''". <<BR>>In the drop-down list, select sensor '''MLP56''' (the one with the strongest respons at 90ms). <<BR>>Right-click on the TF figure > '''Time series'''. <<BR>><<BR>> {{attachment:tf_trials_evaluate.gif||height="163",width="559"}} * The task for defining a baseline is now a lot trickier than before. The epochs are only 600ms long, and the power at many frequencies could not be estimated correctly. If we want all the values to be "good" after 0s, '''we cannot use anything below 15Hz'''. If we want to normalize the values, we have to go even higher: '''30Hz''' '''if we want a baseline of 50ms''' before 0. * The epochs we use in this tutorial are '''too short to perform a correct time-frequency analysis'''. We should have import at least 200ms more on each side, just for controlling the edge effects. You should always think carefully about the length of the epochs you import in your database if you are planning to run any form of frequency or time-frequency analysis. * For the purpose of illustrating the tools available in Brainstorm, we will keep on working with these short epochs, let's try to do the best we can with what we have here. We could use a baseline of 50ms to get a correct estimation above 30Hz, but this is probably a bit too short. We propose to include some more of the baseline (75ms), hoping there are not major edge effects in this segment. * In Process1, select the avereage time-frequency file. * Run process: '''Standardize > Baseline normalization''', Baseline='''[-75, 0]ms''', Method='''ERS/ERD'''. <<BR>><<BR>> {{attachment:tf_trials_normalize.gif||height="285",width="517"}} * The new menus available to display this file are described in the next section. <<BR>><<BR>> {{attachment:tf_trials_popup.gif||height="194",width="625"}} ==== Things to avoid ==== * Avoid computing the time-frequency decomposition of the average, you would miss a lot of the '''induced response''', the brain responses in higher frequencies that are not strictly time-locked to the stimulus, and not aligned in phase across trials. Always prefer the computation of the average of the TF power of each trials, as we did here. Average(TF(trials)), not TF(Average(trials)). <<BR>>This is well documented in: [[http://www.iec-lnc.ens.fr/IMG/Files/Visual/Publications/2000_Review_IntJPsychol.pdf|Bertrand O, Tallon-Baudry C (2000)]]. * Avoid using the "spectral flattening: 1/f compensation" with frequencies above 60 or 80Hz. As mentioned before, it may overcompensate the higher frequencies. If you apply this normalization on the file we just computed, you get figures that give you a false idea of dominant high frequencies: <<BR>><<BR>> {{attachment:tf_trials_multiply.gif||height="129",width="216"}} == Display: All channels == Three menus display the time-frequency of all the sensors with different spatial organizations. All the figures below represent the '''ERS/ERD'''-normalized average. They use the "jet" colormap, which is not the default configuration of this files. To get the same displays, change the colormap configuration: <<BR>>right-click on the figure > Colormap: Stat2 > Colormap > '''jet'''. * '''All channels''': All the maps are displayed one after the other, in the order they are saved in the file. <<BR>><<BR>> {{attachment:tf_allchannels.gif||height="295",width="345"}} * '''2D Layout (maps)''': Show each TF map where the sensor is located on a flattened 2D map. <<BR>><<BR>> {{attachment:tf_2dlayout_maps.gif||height="293",width="345"}} * '''2D Layout (no overlap)''': Similar to the the previous display, but the positions of the images are reorganized so that they do not overlap. <<BR>><<BR>> {{attachment:tf_2dlayout_nooverlap.gif||height="294",width="345"}} Useful shortcuts for these figures: * '''Click''': Clicking on any small TF image opens a new figure with only the selected sensor. * '''Shift + click''': Opens the original recordings time series of the selected sensor, when available. Here, we display an average of time-frequency maps, so this menu has no effect. * '''Mouse wheel''': Zoom in/out. * '''Right click + move''': Move in a zoomed figure. == Display: Topography == The menus below show the distribution of TF power over the sensors, for one time point and one frequency bin, very similarly to what was introduced in tutorial [[Tutorials/ExploreRecordings|Visual exploration]]. * '''2D Sensor cap''' / '''2D Disc''' / '''3D Sensor cap''': 90ms, 8Hz<<BR>><<BR>> {{attachment:tf_topo.gif||height="137",width="474"}} * '''2D Layout''': 8Hz (black), 35Hz (white)<<BR>><<BR>> {{attachment:tf_2dlayout8.gif||height="241",width="260"}} {{attachment:tf_2dlayout35.gif||height="240",width="259"}} Useful shortcuts for these figures: * '''Left/right arrows''': Change the current time. * '''Up/down arrows''': Change the current frequency. * '''Control + E''': Display the sensors markers/names. * '''Shift + click on a sensor''': Displays the time-frequency decomposition for that specific sensors. * '''Right click + move''': Select a group of sensors. * '''Shift +'''''' scroll''': Change the gain of the time series (2D Layout). * '''Control +'''''' scroll''': Change the length of the window displayed around the current time (2D Layout). == Scouts [TODO] == Similar calculations can be done at the level of the sources, either on the full cortex surface or on a limited number of regions of interests. We will start with the latter as it is usually an easier approach.<<BR>>We will use this section to illustrate the online averaging of the trials TF decomposition as well. * Drag and drop all the '''deviant trials''' from '''both runs''', select ['''Process sources''']. * Run process "'''Frequency > Time-frequency (Morlet wavelets)'''". <<BR>>Select the option "Use scouts" and '''select all the scouts''' defined in the previous tutorial. <<BR>><<BR>> {{attachment:scouts_process.gif||height="347",width="448"}} * In the advanced options, select "Scout function: '''After'''" and "Output: '''Save average'''". <<BR>>Run the process (it may take a while).<<BR>><<BR>> {{attachment:scouts_options.gif||height="637",width="380"}} * The scout function was introduced in the previous tutorial. It is the method we use to group the time series for the 40*3=120 unconstrained dipoles we have in each scout into one unique signal per orientation (three signals in the unconstrained case). When computing the TF of one scout, we have the choice between applying this function before or after the time-frequency decomposition itself. * '''Before''': Extract the 120 source signals, apply the scout function to get three signals (one per orientation), run the TF decomposition of the three signals, and finally sum the power of the three TF maps. This is faster but may lose some frequency resolution (especially for constrained sources). * '''After''': Extract the 120 source signals, run the TF decomposition of the 120 signals, apply the scout function on the power of the TF maps for each orientation separately, and finally sum the three orientations. You should always prefer this option if the computation times are not insane. * Rename the new file to add a tag "Deviant" in it. Then right-click > '''Time-freq: All scouts'''. <<BR>><<BR>> {{attachment:scouts_file.gif||height="185",width="508"}} [...] * The normalized maps seem to show a response of the left primary auditory cortex (A1L is the first scout in the file) around 35Hz and 80ms. The large red part at the lower frequency is probably part of the evoked response to the stimulus, but all these values could not be estimated properly because the time window is not long enough. Because we used the baseline between -100ms and 0ms, we cannot really trust anything below 30Hz or 40Hz. * A better approach would have been to import longer epochs (+200ms on each side), then to normalize the values based on the "safe" part of the baseline (100ms-0ms). If you include the edge effects in your baseline, you end up propagating them in all the epoch. * The figures do not use the same colormaps because the original TF file is displayed using the colormap "'''Timefreq'''" while the normalized files are displayed using the colormap "'''Stat 2'''". If you want to look at the normalized maps using a different colormap, right-click on the figure and change the colormap configuration. <<TAG(Advanced)>> == Full cortical maps [TODO] == Computing the time-frequency decomposition for all the sources of the cortex surface is possible but complicated because it can easily generate gigantic files, completely out of the reach of most computers. For instance the full TF matrix for each epoch we have here would be [Nsources x Ntimes x Nfrequencies] = [45000 x 361 x 40] double-complex = '''9.7 Gb'''! We have two ways of going around this issue: computing the TF decomposition for '''fewer frequency bins''' or frequency bands at a time, or computing the '''TF decomposition of the sensor values''' and projecting it dynamically to the cortex using the inverse operator. This latter approach is possible because the two processes (TF decomposition and inversion) are linear and permutable. However this is enabled only for constrained sources (so not possible in the present case). <<BR>>TF(Inverse(Recordings)) = Inverse(TF(Recordings))<<BR>>=> Power(TF(Inverse(Recordings))) = Power(Inverse(TF(Recordings))) For illustration purpose, we will use the other time-frequency decomposition option available in Brainstorm, which is based on Hilbert transforms of band-passed signals, using only 6 predefined frequency bands. * Drag and drop in Process1 the sources for the '''deviant average''' in '''Run#01'''.<<BR>>Select the process "'''Frequency > Hilbert transform'''".<<BR>>To process the entire brain, do '''not '''select the option "Use scouts".<<BR>><<BR>> {{attachment:tf_hilbert_process.gif||height="376",width="480"}} * In the advanced options, select "'''No, save full hilbert(sources)'''" because the other option is not available on unconstrained sources, and "'''magnitude'''" to divide the size of the file by two.<<BR>><<BR>> {{attachment:tf_hilbert_options.gif||height="435",width="374"}} * Right-click on the Hilbert file > '''Display on cortex'''. <<BR>><<BR>> {{attachment:tf_hilbert_popup.gif||height="182",width="284"}} * The frequency slider now shows frequency bands ("alpha:8-12Hz") instead of frequencies ("12Hz").<<BR>>At 90ms, we observe a strong response in the auditory cortex in the alpha band (left) and a response in the motor cortex in the low gamma (right). <<BR>>This could make sense, but this is for illustration purpose only, as we said before we cannot interpret what is happening at 10Hz, and we shouldn't have been computing the decomposition of the average, but rather the average of the TF of all the trials. <<BR>><<BR>> {{attachment:tf_hilbert_alpha.gif||height="175",width="401"}} {{attachment:tf_hilbert_gamma.gif||height="175",width="222"}} * '''Shift + click '''on the cortex surface: Displays the TF decomposition of the selected source.<<BR>>In the TF figure, it doesn't show as a high value because this file hasn't been normalized.<<BR>><<BR>> {{attachment:tf_sources.gif}} * '''Right-click on the brain''': Selects the closest vertex and displays the popup menu at the same time. The first three menus are relative to the source that was just clicked. <<BR>><<BR>> {{attachment:tf_sources_popup.gif}} <<TAG(Advanced)>> == Robustness to noise == Same analysis with no noise: Morlet + <<TAG(Advanced)>> == Getting rid of the edge effects == <<TAG(Advanced)>> == On the hard drive == Right click one of the first TF file we computed > File > '''View file contents'''. . {{attachment:tf_contents.gif||height="450",width="563"}} ==== Structure of the time-frequency files: timefreq_*.mat ==== * '''TF''': [Nsignals x Ntime x Nfreq] matrix containing all the values of the time-frequency decomposition (complex wavelet coefficients, or double values for power/magnitude/Z-score). * '''TFmask''': [Nfreq x Ntime] logical mask indicating the edge effects (0=edge, 1=valid value). * '''Std''': [Nsignals x Ntime x Nfreq] standard deviation if this file is an average. * '''Comment''': String displayed in the database explorer to represent the file. * '''DataType''': From what kind of data this file was computed: {'data', 'results', 'scout, 'matrix'} * '''Time''': [1 x Ntime] Time vector used to estimate this file. * '''TimeBands''': [Ntimebands x 3] Cell array where each line represents a time band: <<BR>>{'band_name', 'time definition', 'function'} * '''Freqs''': For regular frequency binning: vector containing all the frequencies.<<BR>>If using frequency bands: [Nfreqbands x 3] cell array, where each line represents a frequency band {'band_name', 'frequency definition', 'function'} * '''RefRowNames''': Used only for connectivity matrices. * '''RowNames''': [1 x Nsignals] Cell array of strings that describes each row of the TF matrix. In this specific case, it would be the list of all the MEG sensor names. But it could also be a list of names of scouts or clusters. * '''Measure''': Contains the name of the function that was applied right after the computation of the wavelet coefficients. So it represents the type of data contained in the TF matrix. Possible values: * '''none''': No measure applied, TF contains the complex wavelet coefficients. * '''power''': Power for each frequency, ie. the square of the amplitude: abs(coefficients).^2 * '''magnitude''': abs(coefficients) * '''log''': 10 * log10( abs(coefficients).^2) * '''phase''': angle(coefficients) * '''Method''': String that identifies the process that generated the file: <<BR>>{'morlet', 'fft', 'psd', 'hilbert', 'corr', 'cohere', 'granger', 'plv'} * '''DataFile''': Initial file from which this file was computed. In the database explorer, the TF file will be shown as a child of this DataFile file. * '''SurfaceFile''' / '''GridLoc''' / '''GridAtlas''': Source space that was used, only for source files. * '''nAvg''': Number of trials that were averaged to obtain this file. * '''ColormapType''': String, force a specific colormap type to be used when displaying this file. * '''DisplayUnits''': String, force to use specific units when displaying this file. * '''Options''': Options that were selected in the time-frequency options window. * '''History''': List of operations performed on this file (menu File > View file history). ==== Useful functions ==== * '''in_bst_timefreq'''(TimefreqFile): Read a time-frequency file. * '''in_bst'''(FileName, TimeWindow): Read any Brainstorm file with the possibility to load only a specific part of the file. "TimeWindow" is an range of time values in seconds: [tStart, tStop]. * '''bst_process'''(''''LoadInputFile'''', FileName, Target, TimeWindow): The most high-level function for reading data files. "Target" is a string with the list of sensor names or types to load. * '''morlet_transform'''(): Applies complex Morlet wavelet transform to the time series in input. <<TAG(Advanced)>> == References [TODO] == * Bertrand O, Tallon-Baudry C (2000)<<BR>> [[http://www.iec-lnc.ens.fr/IMG/Files/Visual/Publications/2000_Review_IntJPsychol.pdf|Oscillatory gamma activity in humans: a possible role for object representation]]<<BR>>Int J Psychophysiol, 38(3):211-23 * Le Van Quyen M, Foucher J, Lachaux J, Rodriguez E, Lutz A, Martinerie J, Varela FJ (2001) <<BR>>[[http://brainimaging.waisman.wisc.edu/~lutz/LeVanQuyen_et_all_JNM_2001.pdf|Comparison of Hilbert transform and wavelet methods for the analysis of neuronal synchrony]]<<BR>>J Neurosci Methods, 111(2):83-98 * Add reference for bandpass+Hilbert: MX Cohen? <<TAG(Advanced)>> == Additional documentation == * Forum: Time and frequency resolution: http://neuroimage.usc.edu/forums/showthread.php?1848 == Delete all your experiments == Before moving to the next tutorial, '''delete '''all the time-frequency files you computed in this tutorial. It will make the database structure less confusing for the following tutorials. <<HTML(<!-- END-PAGE -->)>> <<EmbedContent("http://neuroimage.usc.edu/bst/get_prevnext.php?prev=Tutorials/Scouts&next=Tutorials/Difference")>> |
Tutorial 24: Time-frequency
[TUTORIAL UNDER DEVELOPMENT: NOT READY FOR PUBLIC USE]
Authors: Francois Tadel, Dimitrios Pantazis, Elizabeth Bock, Sylvain Baillet
This tutorial introduces how to compute time-frequency decomposition of MEG/EEG recordings and cortical currents using complex Morlet wavelets and Hilbert transforms.
Contents
- Introduction
- Morlet wavelets
- Edge effects
- Simulation
- Process options
- Display: Time-frequency map
- Display: Mouse and keyboard shortcuts
- Display: Power spectrum and time series
- Normalized time-frequency maps
- Tuning the wavelet parameters
- Hilbert transform [TODO]
- MEG recordings: Single trials
- Display: All channels
- Display: Topography
- Scouts [TODO]
- Full cortical maps [TODO]
- Robustness to noise
- Getting rid of the edge effects
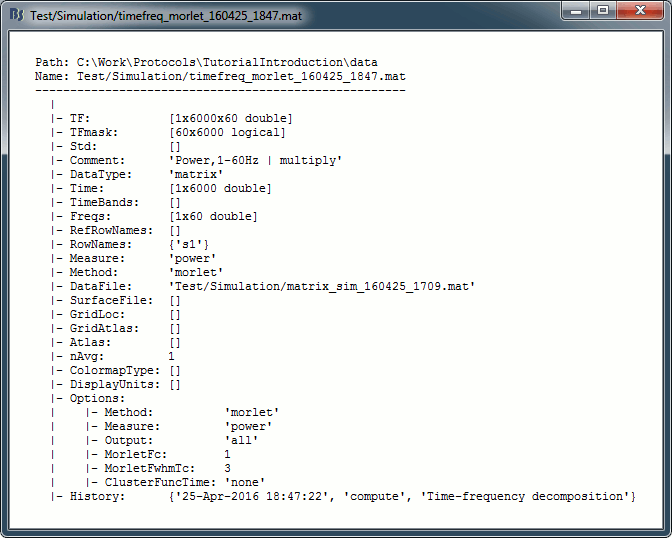
- On the hard drive
- References [TODO]
- Additional documentation
- Delete all your experiments
Introduction
Some of the MEG/EEG signal properties are difficult to access in time domain (graphs time/amplitude). A lot of the information of interest is carried by oscillations at certain frequencies, but the amplitude of these oscillations is sometimes a lot lower than the amplitude of the slower components of the signal, making them difficult to observe.
The averaging in time domain may also lead to a cancellation of these oscillations when they are not strictly locked in phase across trials. Averaging trials in time-frequency domain allows to extract the power of the oscillation regardless of the phase shifts. For a better understanding of this topic, we recommend the lecture of the following article: Bertrand O, Tallon-Baudry C (2000), Oscillatory gamma activity in humans: a possible role for object representation
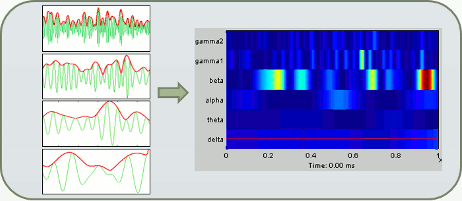
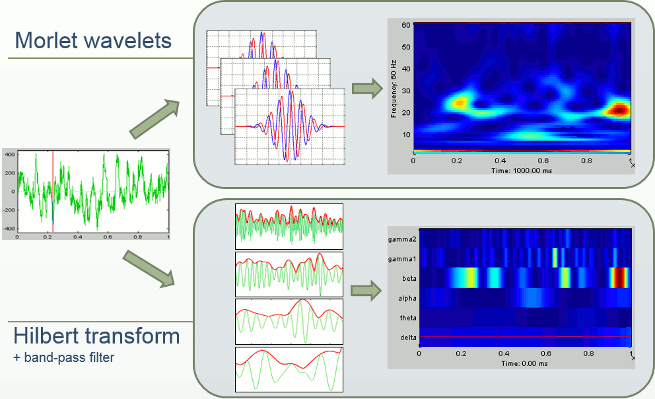
In Brainstorm we offer two approaches for computing time-frequency decomposition (TF): the first one is based on the convolution of the signal with series of complex Morlet wavelets, the second filters the signal in different frequency bands and extracts the envelope of the filtered signals using the Hilbert transform.
Morlet wavelets
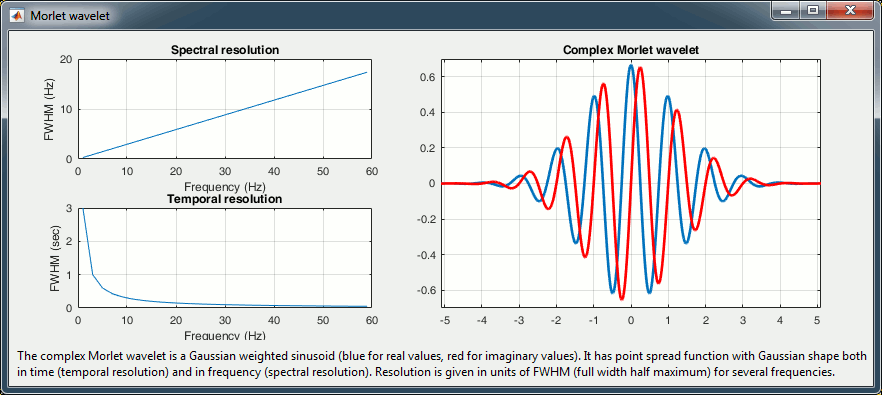
Complex Morlet wavelets are very popular in EEG/MEG data analysis for time-frequency decomposition. They have the shape of a sinusoid, weighted by a Gaussian kernel, and they can therefore capture local oscillatory components in the time series. An example of this wavelet is shown below, where the blue and red curves represent the real and imaginary part, respectively.
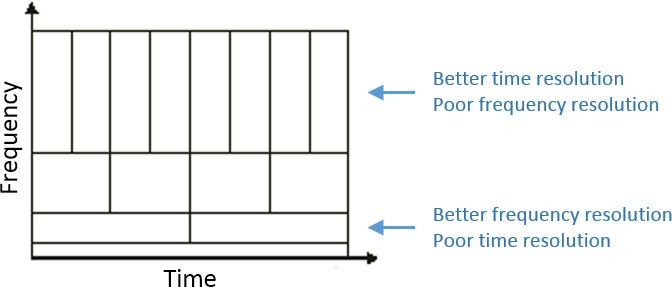
Contrary to the standard short-time Fourier transform, wavelets have variable resolution in time and frequency. For low frequencies, the frequency resolution is high but the time resolution is low. For high frenquies, it's the opposite. When designing the wavelet, we basically decide a trade-off between temporal and spectral resolution.
To design the wavelet, we first need to choose a central frequency, ie. the frequency where we will define the mother wavelet. All other wavelets will be scaled and shifted versions of the mother wavelet. Unless interested in designing the wavelet at a particular frequency band, the default 1Hz should be fine.
Then, the desirable time resolution for the central frequency should be defined. For example, we may wish to have a temporal resolution of 3 seconds at frequency 1 Hz (default parameters). These two parameters, uniquely define the temporal and spectral resolution of the wavelet for all other frequencies, as shown in the plots below. Resolution is given in units of Full Width Half Maximum of the Gaussian kernel, both in time and frequency.
Edge effects
Users should pay attention to edge effects when applying wavelet analysis. Wavelet coefficients are computed by convolving the wavelet kernel with the time series. Similarly to any convolution of signals, there is zero padding at the edges of the time series and therefore the wavelet coefficients are weaker at the beginning and end of the time series.
From the figure above, which designs the Morlet wavelet, we can see that the default wavelet (central frequency fc=1Hz, FWHM_tc=3sec) has temporal resolution 0.6sec at 5Hz and 0.3sec at 10Hz. In such case, the edge effects are roughly half these times: 300ms in 5Hz and 150ms in 10Hz.
More precisely, if f is your frequency of interest, you can expect the edge effects to span over FWHM_t seconds: FWHM_t = FWHM_tc * fc / f / 2. Examples of such transients are given in the figures below.
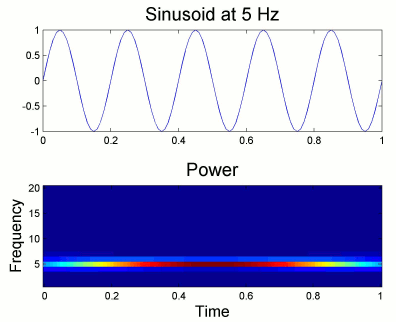
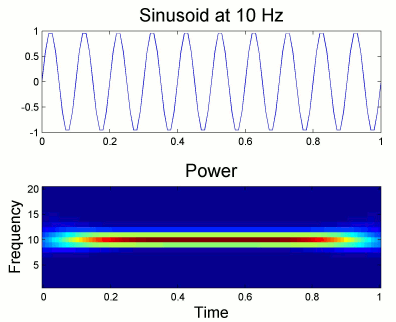
Simulation
We will illustrate the time-frequency decomposition process with a simulated signal.
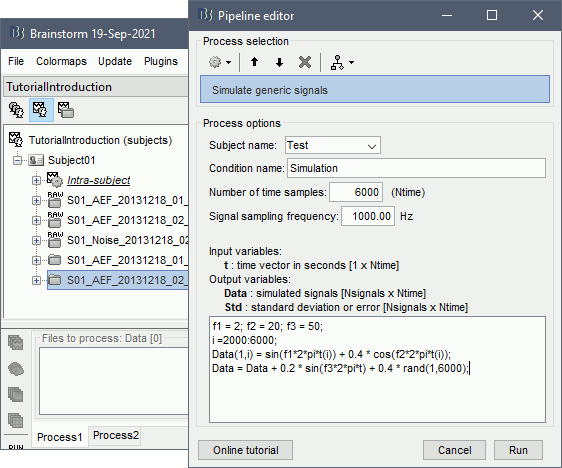
The following code generates a sum of three sinusoids (2Hz, 20Hz, 50Hz) with random white noise. The 50Hz and noise are present everywhere, the 2Hz and 20Hz start only after two seconds.
f1 = 2; f2 = 20; f3 = 50; i =2000:6000; Data(1,i) = sin(f1*2*pi*t(i)) + 0.4 * cos(f2*2*pi*t(i)); Data = Data + 0.2 * sin(f3*2*pi*t) + 0.4 * rand(1,6000);
Empty the Process1 list (right-click > Clear list) then click on [Run].
Run process: Simulate > Simulate generic signals.
Ntime=6000, Sampling frequency=1000Hz (signal duration = 6000/1000 = 6 seconds).
Copy-paste the few lines of code above to generate the sum of three sinusoids.
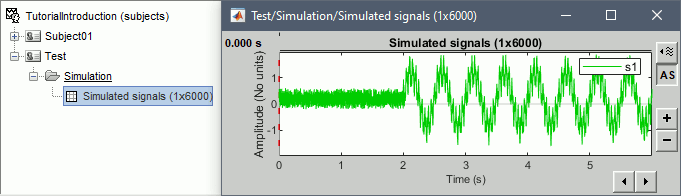
Double-click on the new file to look at the simulated signal.
- In Process1, select the simulated signal.
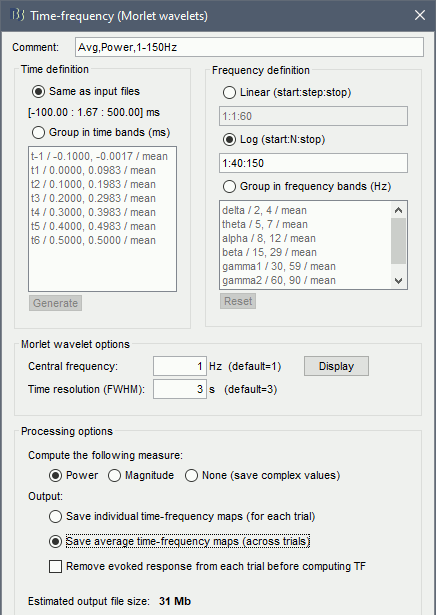
Run process: Frequency > Time-frequency (Morlet wavelets).
Select the option 1/f compensation: The normalization will be discussed later.
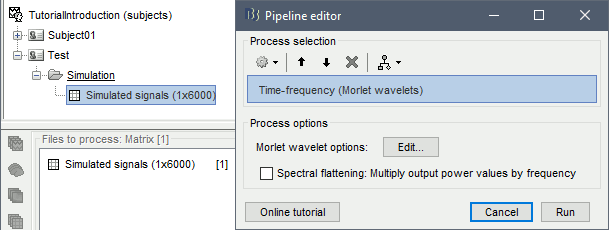
Click on the button [Edit] to see all the process options.
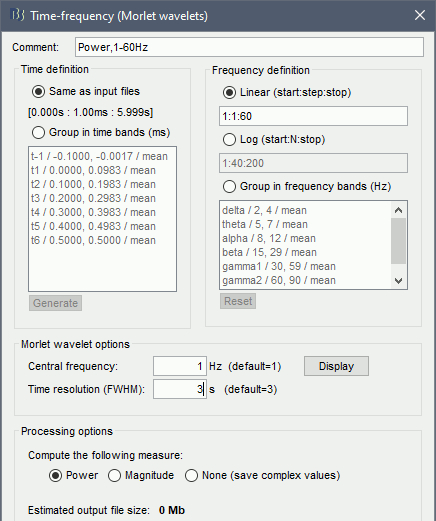
Time definition: Same as input, Frequency definition: Linear 1:1:60, Compute measure: Power.
Process options
Comment: String that will be displayed in the database explorer to represent the output file.
Time definition
Same as input file: The output file has the same time definition as the input file.
In this example, it means: 6000 samples between 0 and 6s.Group in time bands: This option adds a step of computation. First it computes the TF decomposition for all the input file, then averages the power by time band. To define a time band:
Enter your own time bands in the text area, one line per time band, with the following format: "name / time definition / function"
- Click on the button [Generate] to automatically create a list of time bands with the same length. You will be asked the maximal length of each time band.
The function is the measure we take to combine the values for all the individual frequencies into one for the frequency band. Possible values are: mean, max, std, median.
Frequency definition: Frequencies for which the power will be estimated at each time instant.
Linear: You can specify the frequencies with the Matlab syntax start:step:stop.
The default is "1:1:60", which produces 60 values [1, 2, 3, 4, ..., 59, 60].Log: With the option start:N:stop, produces a list of N frequencies logarithmically scaled between "start" and "stop". For example "1:40:80" is converted to [1, 1.5, 2.1, 2.7, ..., 61.5, 65.8, 75, 80]
Group in frequency bands: As for the time definition, this option leads to a two-step process. First it computes the TF decomposition for several values in the frequency band, then it averages the power of TF coefficients per frequency band. To define a frequency band:
One line per frequency band, with the format "name / frequency definition / function"
The frequency definition is a Matlab expression evaluated with an eval() call. If the frequency definition contains only two values, Brainstorm adds two extra values in the middle so that the final averaged value is a bit more robust. Example of valid expressions:
"2,4": Evaluates to [2,4], and then expands to the frequency vector [2, 2.66, 3.33, 4]
"2:0.5:4": Evaluates to [2 2.5 3 3.5 4]
"2, 2.5, 3, 3.5, 4": Evaluates to [2 2.5 3 3.5 4]The function is the measure we take to combine the values for all the individual frequencies into one for the frequency band. Possible values are: mean, max, std, median.
Morlet wavelet options
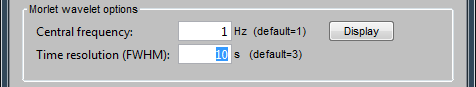
Central frequency: Frequency where the mother wavelet is designed. All other wavelets will be shifted and scaled versions of the mother wavelet
Time resolution (FWHM): Temporal resolution of the wavelet at the central frequency (in units of Full Width Half Maximum). Click [Display] to see the resolution of the wavelet for other frequencies.
Compute the following measure:
- The convolution of the signal with complex Morlet wavelets returns the complex coefficients for each frequency/time/sensor. Typically, what we display is the power of the coefficients (square of the amplitude: abs(TF).^2). You can choose if you want to apply this transformation or not.
Power: Computes the "power" transformation immediately after the TF decomposition. This discards the phase information, but produces files that are twice smaller and a lot easier to process.
None: Save the TF coefficients as they are computed. This can be useful if you plan to use these decompositions for other purposes that require the phase.
- Some combinations of options may disable this choice. If you select frequency bands, the program will have to compute the power before averaging the values, therefore "none" is not an option.
Display: Time-frequency map
Right on the new time-frequency file > Time-freq: One matrix (same as double-clicking).
This menu displays the time-frequency decomposition of the first (and only) signal. The Brainstorm window shows two new elements: the tab "Display" and the frequency slider.
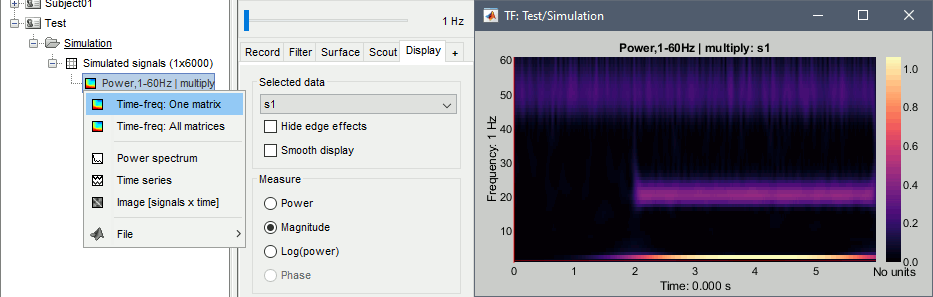
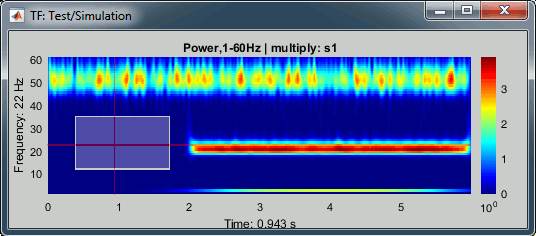
- We can easily identify the three horizontal bars as the three sinusoids in the simulated signals, and the trade-off between accuracy in time and accuracy in frequency. Click on the figure to move the time-frequency cursor and explore the two axes of the plane.
2Hz: High frequency resolution but poor time resolution (supposed to start sharply at 2s)
20Hz: Better time resolution but poorer frequency resolution (17-24Hz)
50Hz: Continuous over the 6s - Frequency resolution gets even worse (40-60Hz). It looks discontinuous because this oscillation has the same amplitude as the white noise we added in the signal (weight 0.4, relatively to the 2Hz oscillation).
Current frequency: Slider that shows the current frequency selected in all the figures.
Just like the time, the frequency selection is centralized and managed by one control only for all the figures. As a consequence, it is impossible to display TF files with different frequency definitions at the same time. This can be perceived as an annoying limitation, but it allows all the simultaneous displays to be consistent at anytime and makes the interface more intuitive to manipulate, with lower risks of mistakes in the interpretation of the different figures.List of signals: This drop-down list shows the signal currently displayed in the selected figure. In this case, there is only one channel of data called "s1". It will be more useful later.
Hide edge effects: When this option is selected, the time-frequency coefficients that could not be properly estimated because of a lack of time samples are hidden. It allows you to see only the information that is really reliable. The lower the frequency, the longer the edge effects.
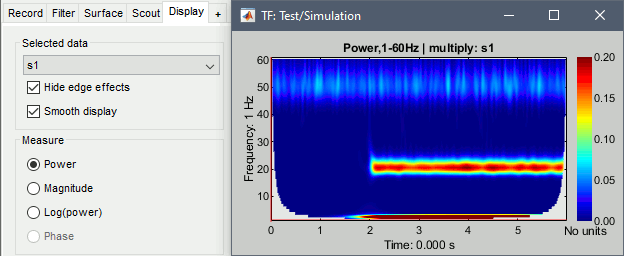
Smooth display: Re-interpolates the time-frequency maps on a finer grid to produce nicer plots.
Measure: Type of measure that is currently represented in the selected figure. The entries that are enabled depend on the type of data that is saved in the file. In this case, we saved directly the power of the wavelet coefficients in the file, we discarded the angle/phase information, so the "phase" option is disabled. The other options are: Magnitude = sqrt(power), Log = 10*log10(power)
Colormap: As explained in the previous tutorials, you can change the colormap by clicking+moving on the colorbar on the right of the figure. Double-click on the colorbar to restore the defaults.
Display: Mouse and keyboard shortcuts
Mouse shortcuts
Left-click: Selection of current time and frequency.
Left-click + move: Select a time/frequency range. The legends of the X/Y axis show the selection.
Mouse wheel: Zoom in time, centered on the current time cursor.
Control + mouse wheel: Zoom in frequencies, centered on the current frequency cursor.
Right-click + move, or Control + left-click + move: Move in the zoomed image.
Double-click: Restore initial view.
Keyboard shortcuts:
Left/right arrows: Change the current time.
Page-up/page-down: Change the current time, 10 time samples at a time.
Up/down arrows: Change the the sensor displayed in this figure.
Control + up/down arrows: Change the current frequency.
Enter: View the original time series for this sensor.
Control + R: View the original MEG recordings.
Control + T: View the time-frequency 2D topography.
Control + I: Save as image.
Control + D: Dock figure in the Matlab environment.
Figure popup menu
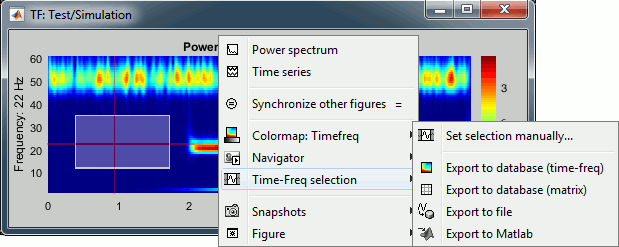
Set selection manually: Does the same thing as drawing a time/freq selection square on a figure, but by typing the values for time and frequency manually.
Export to database: Save the selection for the displayed sensor in a new file in the database.
Export to file: Same as "Export to database", but the saved file is not registered in the database.
Export to Matlab: Same as "Export to database", but the output structure is sent to a variable in the Matlab base workspace instead of being saved to a file.
Display: Power spectrum and time series
These two menus generate similar figures:
Power spectrum: For the current time, shows the power for all the frequencies.
Time series: For the current frequency, shows the power for all the time samples.
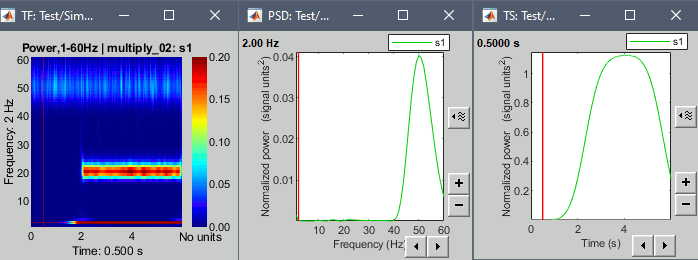
Example: Power spectrum density at 0.5s and power time series at 2Hz.
We see the oscillation at the 50Hz in the PSD plot, and the oscillation at 2Hz in the TS plot.
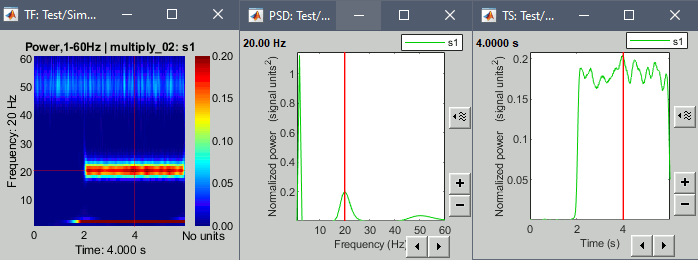
Example: Power spectrum density at 4s and power time series at 20Hz.
We see all three oscillations in the PSD plot, and the oscillation at 20Hz in the TS plot.
Note that if you right-click on the file in the database explorer and then select one of these menus, it will show all the signals. If you right-click on an existing time-frequency figure, it will show only the selected signal. It doesn't make any difference here because there is only one signal, but it will with the MEG recordings.
Normalized time-frequency maps
The brain is always active, the MEG/EEG recordings are never flat, some oscillations are always present in the signals. Therefore we are often more interested in the transient changes in the power at certain frequencies than at the actual power. A good way to observe these changes is to compute a deviation of the power with respect with a baseline.
There is another reason for which we are usually interested in standardizing the TF values. The power of the time-frequency coefficients are always lower in the higher frequencies than in the lower frequencies, the signal carries a lot less power in the faster oscillations than in the slow brain responses. This 1/f decrease in power is an observation that we already made with the power spectrum density in the filtering tutorial. If we represent the TF maps with a linear color scale, we will always see values close to zero in the higher frequency ranges. Normalizing each frequency separately with respect with a baseline helps obtaining more readable TF maps.
No normalization
The values we were looking at were already normalized (option: "1/f compensation" in the process options), but not with respect to a baseline. We will now compute the non-normalized power obtained with the Morlet wavelets and try the various options available for normalizing them.
- In Process1, keep the simulated signal selected.
Run process: Frequency > Time-frequency (Morlet wavelets), "Spectral flattening: None"
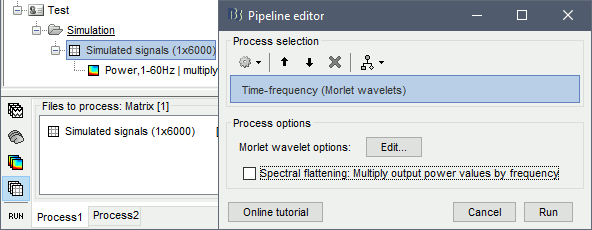
Double-click on the file. As expected, we only see the lower frequencies in this representation: the power of the 2Hz oscillation is a lot larger than the power at 20Hz or 60Hz.
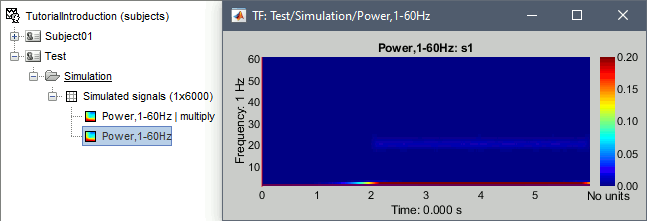
Spectral flattening
- In Process1: Select this new non-normalized file "Power,1-60Hz".
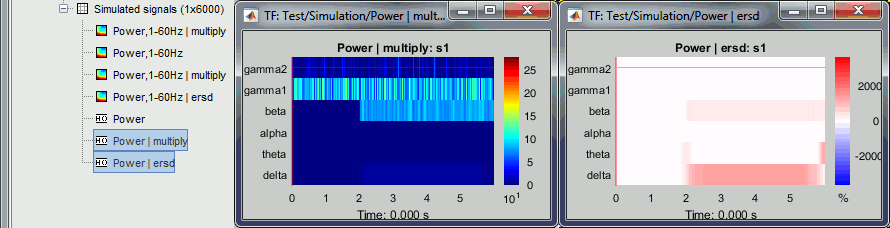
Run process: Standardize > Spectral flattening, Method=1/f compensation.
This produces exactly the same results as previously (option "1/f compensation" in the time-frequency process). It multiplies the power at each frequency bin with the frequency value (eg. multiplies by 20 the power at 20Hz), in order to correct for the 1/f shape we observe in the power spectrum. This works well for the lower part of the spectrum and up to 60-80Hz, but past this range it tends to overcompensate the higher frequencies.
Note that it does not do any form of baseline correction: the 50Hz oscillation is visible everywhere.
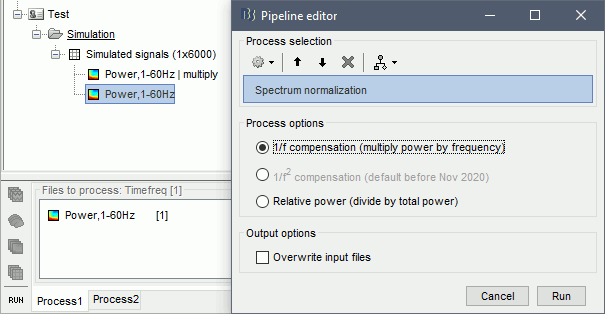
Baseline normalization
The second way to proceed is to normalize the power with respect to its average level during a reference time period. We can consider the oscillations at 2Hz and 20Hz as our events of interest, and the 50Hz as noise we want to get rid of. The segment from 0 to 2 seconds does not contain any of the signals of interest, therefore we can consider it as a baseline.
However, we will not be able to use the full segment [0,2]s because of the edge effects we described at the beginning of this tutorial. The time-frequency map at 2Hz with the display option "Hide edge effects" (left figure below) shows that the power could not be estimated correctly before 0.75s, therefore we shouldn't use it as a baseline. The power time series at 2Hz (right) shows that the power related with the 2Hz oscillation starts to increase significantly after 1.25s, therefore it's not really a "baseline" anymore. This leaves only the segment [0.75,1.25]s available.
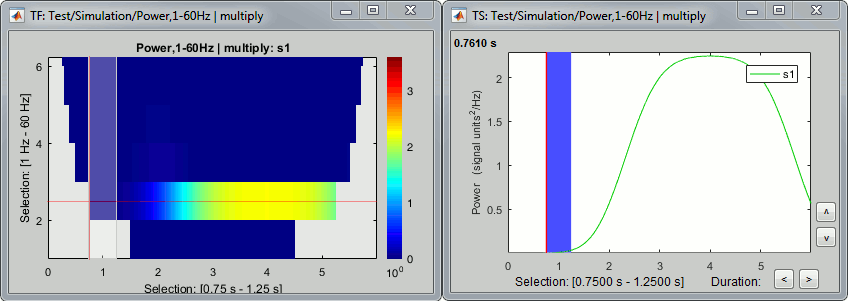
At 20Hz, the expected transient effects are only 75ms long, therefore we could use a much longer baseline if we were not interested by the lower frequencies: [0.075, 1.925]s.
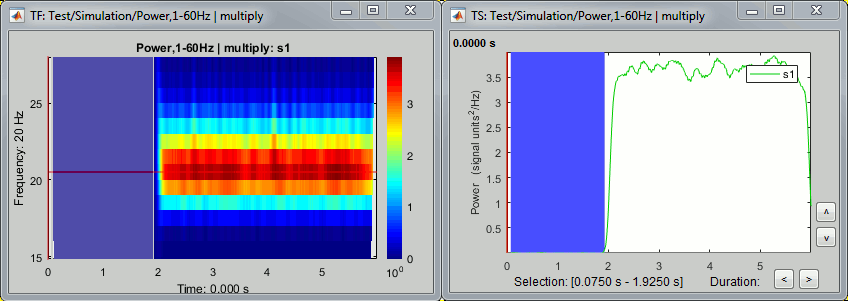
In this case, we have a very long "resting" time segment (2s), therefore the edge effects are not a problem for picking a time window for the baseline normalization. We will use the first time window mentioned, [0.75,1.25]s as it is long enough (500ms) to estimate the baseline mean power. In real-life cases, with shorter epochs, it is sometimes difficult to find an acceptable trade-off between the baseline duration and the exclusion of the edge effects, especially for the lower frequencies.
Run process: Standardize > Baseline normalization, Baseline=[0.75, 1.25]s.
Method=Event-related perturbation: ERS/ERD stands for "event-related synchronization / desynchronization", a widely used normalization measure for time-frequency power maps. It evaluates the deviation from the mean over the baseline, in percents: (x-mean)/mean*100.
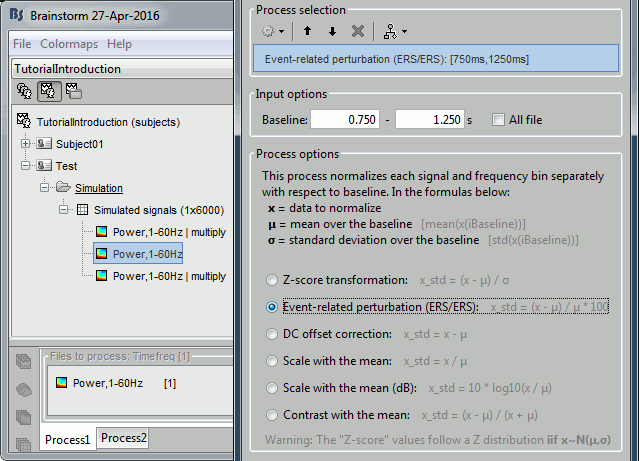
Double-click on the new "ersd" file. The colormap type changed from "Timefreq" to "Stat2", which uses by default the "rwb" color set and shows relative values. Indeed, the ERS/ERD values can be positive or negative: the power at a given time sample can be higher or lower than the average level during the baseline. In the simple simulation we used, there is no power decrease at any frequency after 2s, so the strong values are mostly positive. However, if you look in the file, you would see that there are many small negative values (due to the random noise we added).
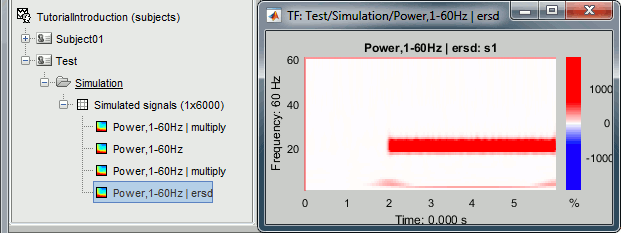
Note that the 50Hz disappeared because it was present during the baseline, while the 2Hz and 20Hz oscillations show high positive values (between 100% and 2000% increase from baseline).
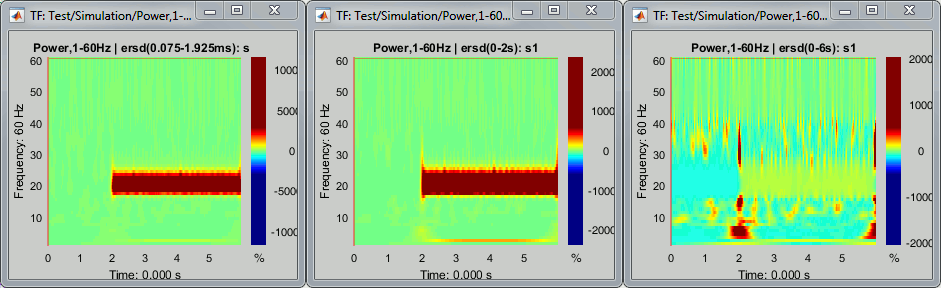
Remember to select your baseline very carefully according to the frequency range you are interested in. See below examples obtained with different baselines: 0.075-1.925s, 0-2s, 0-6s.
Change the colormap to "jet" if you prefer, and adjust the colormap contrast as needed.
Tuning the wavelet parameters
Time resolution
You can adjust the relative time and frequency resolution of your wavelet transformation by adjusting the parameters of the mother wavelet in the options of the process.
Increasing the option "time resolution" will produce longer wavelets at a given frequency, hence increase the frequency accuracy (lower Δf) and decrease the time accuracy (higher Δt). Expect longer edge effects.
Decrease the time resolution will produce shorter wavelets at a given frequency, hence decrease the frequency accuracy (higher Δf) and increase the time accuracy (higher Δt). Expect shorter edge effects.
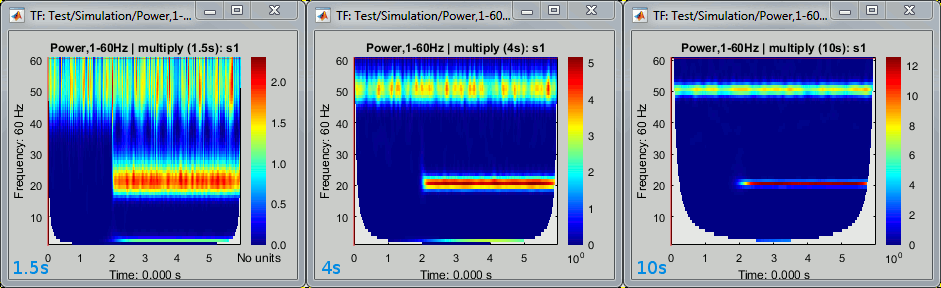
You can modify one or the other parameter, what is important is the product of the two. All the following combinations fc/FWHM_t produce the same results because their product is constant: (1Hz,3s), (3Hz,1s), (6Hz,0.5s), (60Hz,0.05s)
Examples for a constant central frequency of 1Hz with various time resolutions: 1.5s, 4s, 10s.
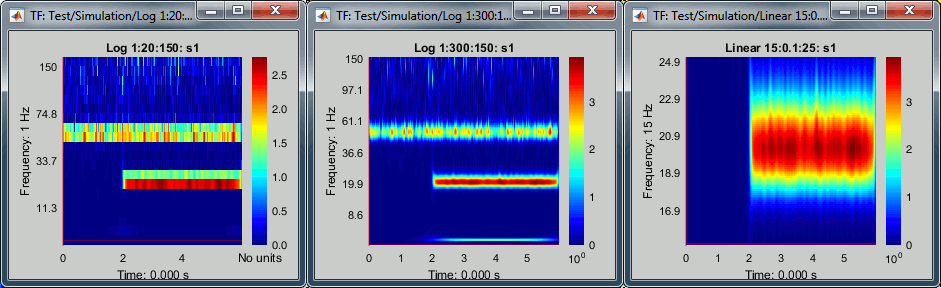
Frequency axis
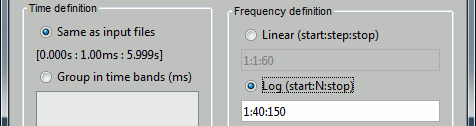
You can also obtain very different representations of the data by changing the list of frequencies for which you estimate the power. You can also change this in the options of the process.
Examples: Log 1:20:150, Log 1:300:150, Linear 15:0.1:25
Hilbert transform [TODO]
We can repeat the same analysis with the other approach available for exploring the simulated signal in the time-frequency plane. The process "Frequency > Hilbert transform" first filters the signals in various frequency bands with a band-pass filter, then computes the Hilbert transform of the filtered signal. The magnitude of the Hilbert transform of a narrow-band signal is a measure of the envelope of this signal, and therefore gives an indication of the activity in this frequency band.
No normalization
Let's compute the same three results as before: non-normalized, 1/f compensation, baseline normalization.
- In Process1, select the simulated signal.
Run process: Frequency > Hilbert transform, Spetral flattening=None, Mirror signal before.
The mirroring option helps with controlling the amplitude of the edge effects.
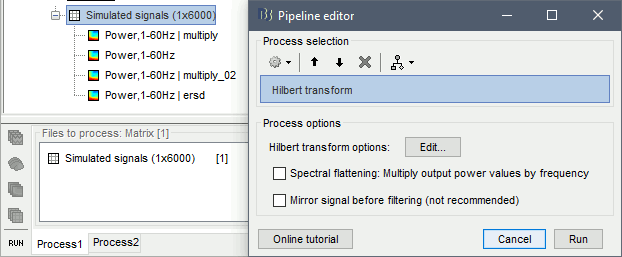
In the advanced options panel, keep the default options: Default frequency bands and Magnitude.
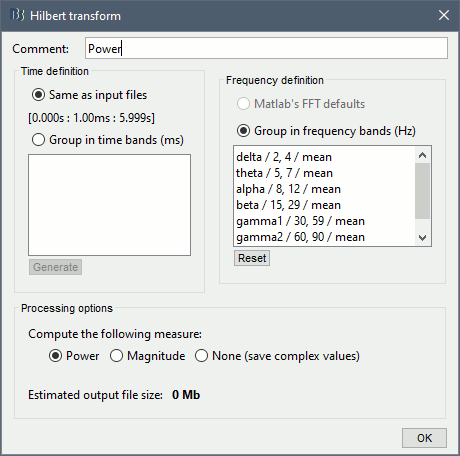
Double-click on the new file. The figure now has only 6 rows, one for each frequency band.
Note that the values saved in the file and displayed are now amplitude and not power anymore.
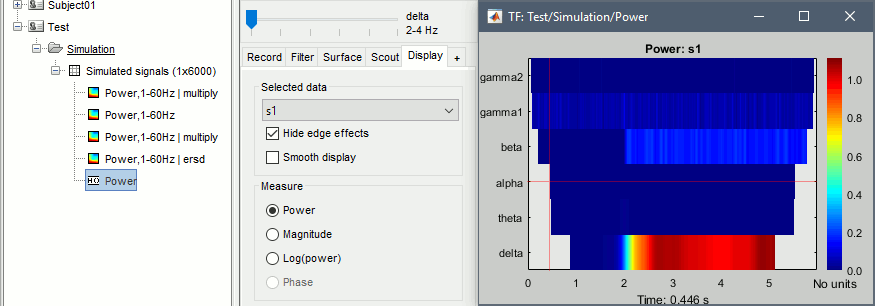
- The non-normalized results are already easy to interpret:
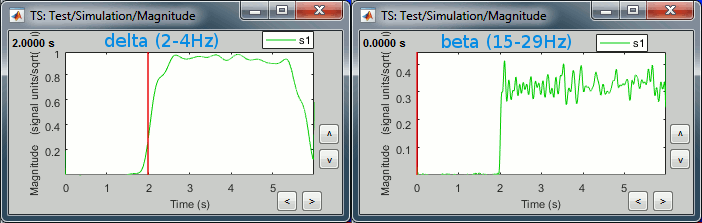
delta (2-4Hz): Includes the 2Hz oscillation, contribution starts at 2s
beta (15-29Hz): Includes the 20Hz oscillation, contribution starts at 2s
gamma1(30-59Hz): Includes the 50Hz oscillation, contribution starts at the beginning (0s)
Right-click on the file or figure > Time series. Example for delta and beta.
Normalization
- In Process1, select the non-normalized Hilbert-based decomposition.
Run process: Standardize > Spectral flattening, Method=1/f compensation.
Run process: Standardize > Baseline normalization, Baseline=[0.75, 1.25]s, Method=ERS/ERD.
Display the two normalized files side by side.
Method specifications
Filters: [TODO]
Hilbert transformation: Using the Matlab's hilbert() function.
- Extraction of the envelope: Magnitude of the complex Hilbert transformat, abs(hilbert(x)).
Magnitude vs power
Be careful with the direct comparison you obtain between this approach and the previous one. By default, the Hilbert process saves everything in amplitude (magnitude of the Hilbert transform) while the Morlet wavelet process saves everything in power (square of the wavelet coefficients).
- This follows the common practices in two different fields of signal processing: envelopes are typically displayed in amplitude, Fourier-transform and other spectral decompositions are displayed in power.
- So even if it looks like these two processes produce similar outputs, they don't. Make sure you take into account this difference in your statistical analysis and reports.
MEG recordings: Single trials
Now let's go back to our auditory oddball paradigm and apply the concepts to MEG recordings. We will use all the trials available for one condition to estimate the average time-frequency decomposition.
Spectral flattening
In Process1, select all the deviant trials in Run#01.
Run process: Frequency > Time-frequency (Morlet wavelets), Spectral flattening: None.
In the advanced options, select: Log 1:40:150, Power, Save average time-frequency maps.
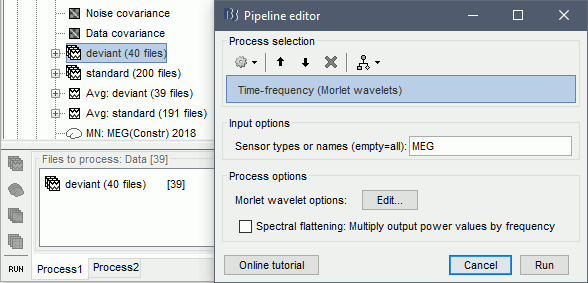
Save individual TF maps: This option stops the computation here and saves in the database one time-frequency file for each input file (40 files), with one TF map for each scout.
Save average TF maps: Instead of saving the TF for each file separately, it automatically computes the average of the power of all the TF. This is a good choice if you do not plan to use independently all the TF files, because it saves a lot of time and disk space.
Remove evoked response from each trial before computing TF: This option first computes the average of all the trials, then subtracts this average from each trial before computing the time-frequency decomposition. This brings the signals to a slightly more stationnary state, which may help for the evaluation of the frequency contents.
Baseline normalization
Double-click on the new file Avg,Power,1-150Hz (MEG). Select "Hide edge effects".
In the drop-down list, select sensor MLP56 (the one with the strongest respons at 90ms).
Right-click on the TF figure > Time series.
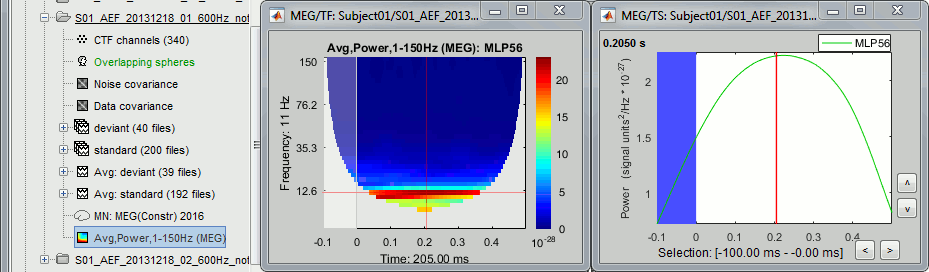
The task for defining a baseline is now a lot trickier than before. The epochs are only 600ms long, and the power at many frequencies could not be estimated correctly. If we want all the values to be "good" after 0s, we cannot use anything below 15Hz. If we want to normalize the values, we have to go even higher: 30Hz if we want a baseline of 50ms before 0.
The epochs we use in this tutorial are too short to perform a correct time-frequency analysis. We should have import at least 200ms more on each side, just for controlling the edge effects. You should always think carefully about the length of the epochs you import in your database if you are planning to run any form of frequency or time-frequency analysis.
- For the purpose of illustrating the tools available in Brainstorm, we will keep on working with these short epochs, let's try to do the best we can with what we have here. We could use a baseline of 50ms to get a correct estimation above 30Hz, but this is probably a bit too short. We propose to include some more of the baseline (75ms), hoping there are not major edge effects in this segment.
- In Process1, select the avereage time-frequency file.
Run process: Standardize > Baseline normalization, Baseline=[-75, 0]ms, Method=ERS/ERD.
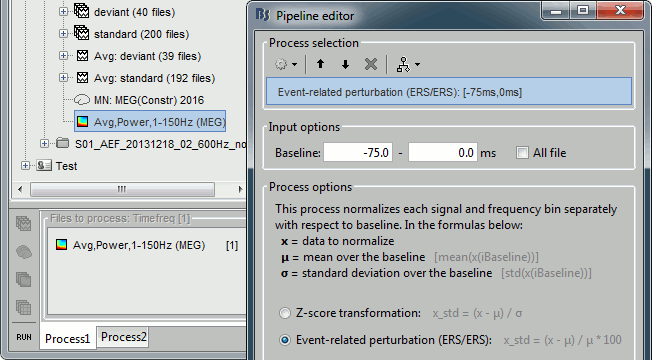
The new menus available to display this file are described in the next section.
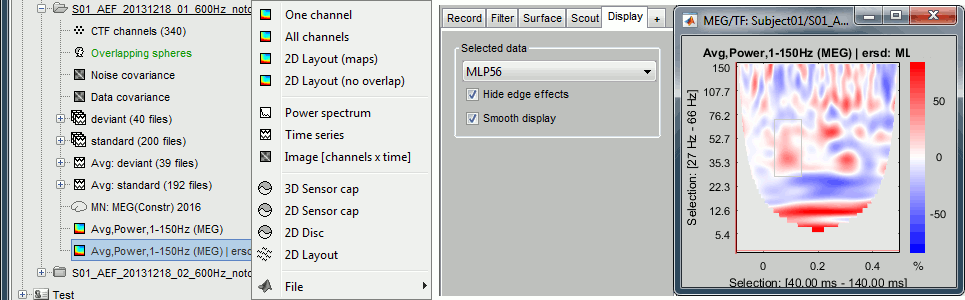
Things to avoid
Avoid computing the time-frequency decomposition of the average, you would miss a lot of the induced response, the brain responses in higher frequencies that are not strictly time-locked to the stimulus, and not aligned in phase across trials. Always prefer the computation of the average of the TF power of each trials, as we did here. Average(TF(trials)), not TF(Average(trials)).
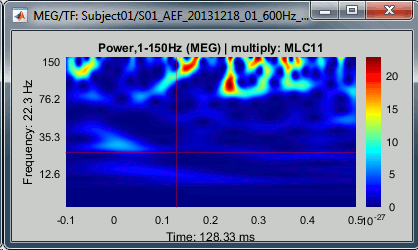
This is well documented in: Bertrand O, Tallon-Baudry C (2000).Avoid using the "spectral flattening: 1/f compensation" with frequencies above 60 or 80Hz. As mentioned before, it may overcompensate the higher frequencies. If you apply this normalization on the file we just computed, you get figures that give you a false idea of dominant high frequencies:
Display: All channels
Three menus display the time-frequency of all the sensors with different spatial organizations. All the figures below represent the ERS/ERD-normalized average. They use the "jet" colormap, which is not the default configuration of this files. To get the same displays, change the colormap configuration:
right-click on the figure > Colormap: Stat2 > Colormap > jet.
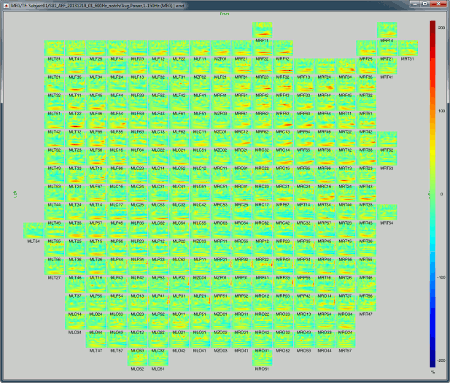
All channels: All the maps are displayed one after the other, in the order they are saved in the file.
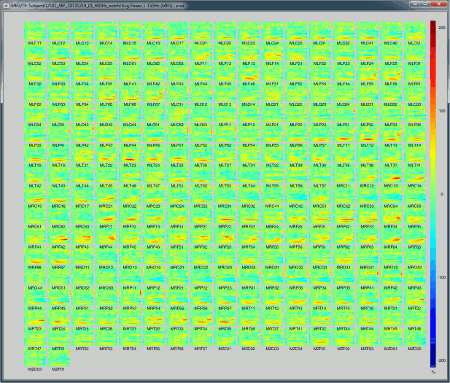
2D Layout (maps): Show each TF map where the sensor is located on a flattened 2D map.
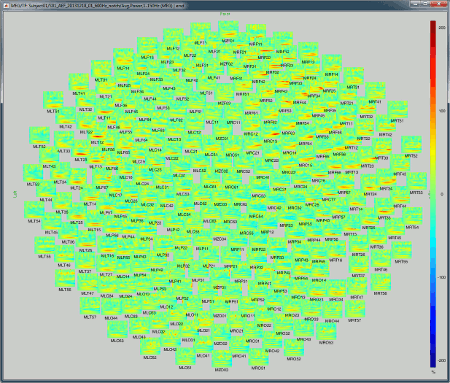
2D Layout (no overlap): Similar to the the previous display, but the positions of the images are reorganized so that they do not overlap.
Useful shortcuts for these figures:
Click: Clicking on any small TF image opens a new figure with only the selected sensor.
Shift + click: Opens the original recordings time series of the selected sensor, when available. Here, we display an average of time-frequency maps, so this menu has no effect.
Mouse wheel: Zoom in/out.
Right click + move: Move in a zoomed figure.
Display: Topography
The menus below show the distribution of TF power over the sensors, for one time point and one frequency bin, very similarly to what was introduced in tutorial Visual exploration.
2D Sensor cap / 2D Disc / 3D Sensor cap: 90ms, 8Hz
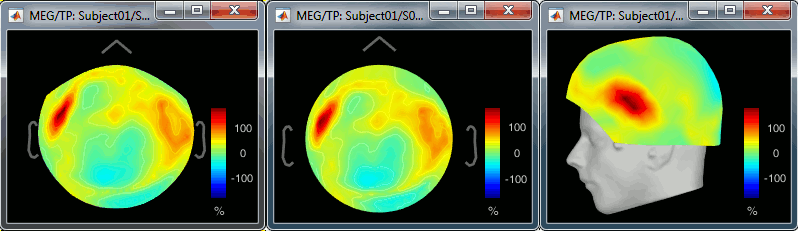
2D Layout: 8Hz (black), 35Hz (white)
Useful shortcuts for these figures:
Left/right arrows: Change the current time.
Up/down arrows: Change the current frequency.
Control + E: Display the sensors markers/names.
Shift + click on a sensor: Displays the time-frequency decomposition for that specific sensors.
Right click + move: Select a group of sensors.
Shift + scroll: Change the gain of the time series (2D Layout).
Control + scroll: Change the length of the window displayed around the current time (2D Layout).
Scouts [TODO]
Similar calculations can be done at the level of the sources, either on the full cortex surface or on a limited number of regions of interests. We will start with the latter as it is usually an easier approach.
We will use this section to illustrate the online averaging of the trials TF decomposition as well.
Drag and drop all the deviant trials from both runs, select [Process sources].
Run process "Frequency > Time-frequency (Morlet wavelets)".
Select the option "Use scouts" and select all the scouts defined in the previous tutorial.
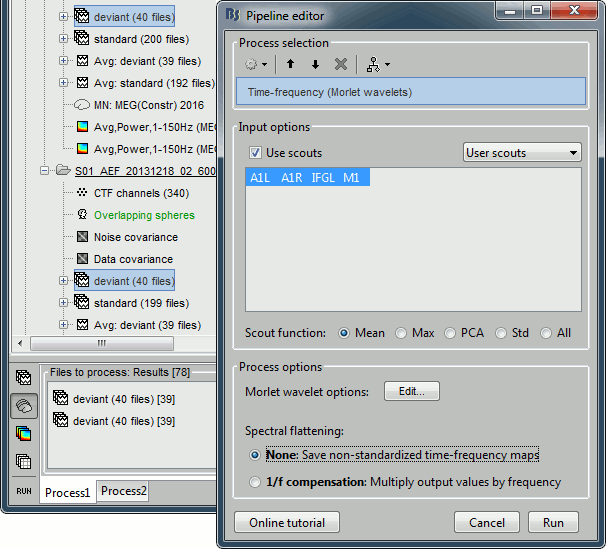
In the advanced options, select "Scout function: After" and "Output: Save average".
Run the process (it may take a while).
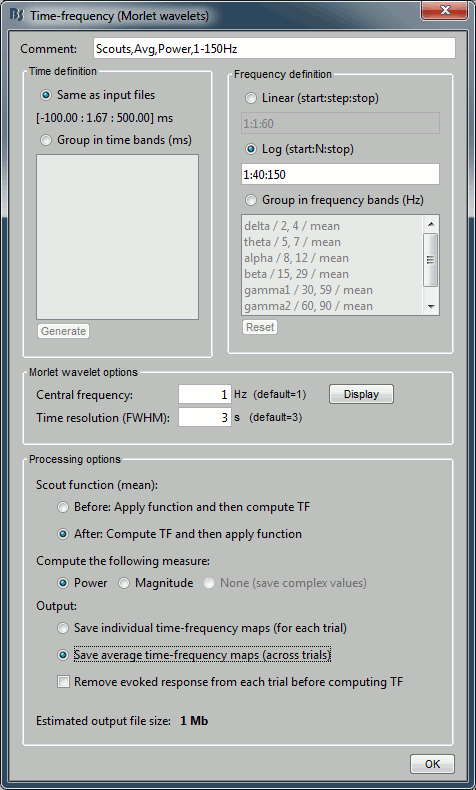
- The scout function was introduced in the previous tutorial. It is the method we use to group the time series for the 40*3=120 unconstrained dipoles we have in each scout into one unique signal per orientation (three signals in the unconstrained case). When computing the TF of one scout, we have the choice between applying this function before or after the time-frequency decomposition itself.
Before: Extract the 120 source signals, apply the scout function to get three signals (one per orientation), run the TF decomposition of the three signals, and finally sum the power of the three TF maps. This is faster but may lose some frequency resolution (especially for constrained sources).
After: Extract the 120 source signals, run the TF decomposition of the 120 signals, apply the scout function on the power of the TF maps for each orientation separately, and finally sum the three orientations. You should always prefer this option if the computation times are not insane.
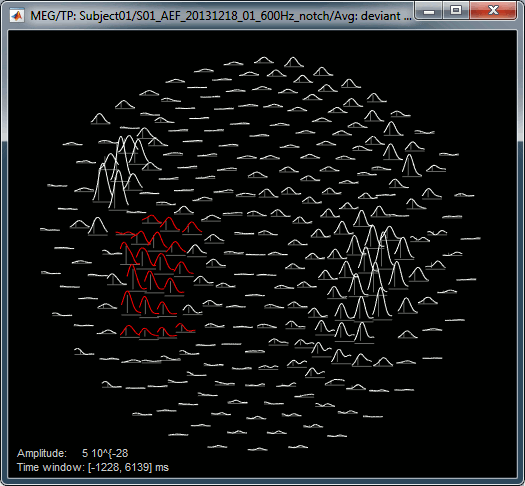
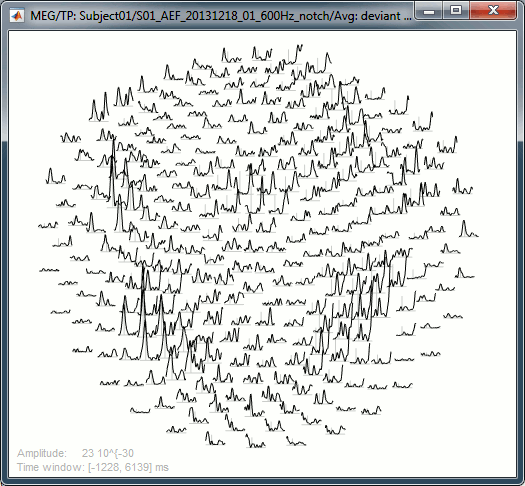
Rename the new file to add a tag "Deviant" in it. Then right-click > Time-freq: All scouts.
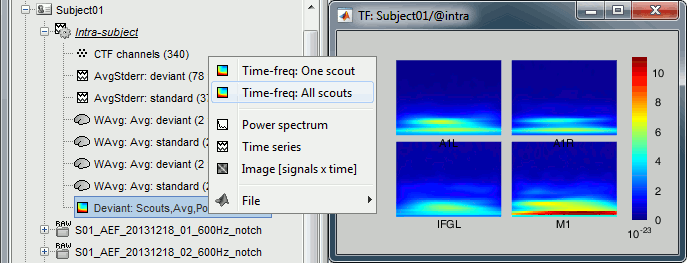
[...]
- The normalized maps seem to show a response of the left primary auditory cortex (A1L is the first scout in the file) around 35Hz and 80ms. The large red part at the lower frequency is probably part of the evoked response to the stimulus, but all these values could not be estimated properly because the time window is not long enough. Because we used the baseline between -100ms and 0ms, we cannot really trust anything below 30Hz or 40Hz.
- A better approach would have been to import longer epochs (+200ms on each side), then to normalize the values based on the "safe" part of the baseline (100ms-0ms). If you include the edge effects in your baseline, you end up propagating them in all the epoch.
The figures do not use the same colormaps because the original TF file is displayed using the colormap "Timefreq" while the normalized files are displayed using the colormap "Stat 2". If you want to look at the normalized maps using a different colormap, right-click on the figure and change the colormap configuration.
Full cortical maps [TODO]
Computing the time-frequency decomposition for all the sources of the cortex surface is possible but complicated because it can easily generate gigantic files, completely out of the reach of most computers. For instance the full TF matrix for each epoch we have here would be [Nsources x Ntimes x Nfrequencies] = [45000 x 361 x 40] double-complex = 9.7 Gb!
We have two ways of going around this issue: computing the TF decomposition for fewer frequency bins or frequency bands at a time, or computing the TF decomposition of the sensor values and projecting it dynamically to the cortex using the inverse operator.
This latter approach is possible because the two processes (TF decomposition and inversion) are linear and permutable. However this is enabled only for constrained sources (so not possible in the present case).
TF(Inverse(Recordings)) = Inverse(TF(Recordings))
=> Power(TF(Inverse(Recordings))) = Power(Inverse(TF(Recordings)))
For illustration purpose, we will use the other time-frequency decomposition option available in Brainstorm, which is based on Hilbert transforms of band-passed signals, using only 6 predefined frequency bands.
Drag and drop in Process1 the sources for the deviant average in Run#01.
Select the process "Frequency > Hilbert transform".
To process the entire brain, do not select the option "Use scouts".
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
In the advanced options, select "No, save full hilbert(sources)" because the other option is not available on unconstrained sources, and "magnitude" to divide the size of the file by two.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
The frequency slider now shows frequency bands ("alpha:8-12Hz") instead of frequencies ("12Hz").
At 90ms, we observe a strong response in the auditory cortex in the alpha band (left) and a response in the motor cortex in the low gamma (right).
This could make sense, but this is for illustration purpose only, as we said before we cannot interpret what is happening at 10Hz, and we shouldn't have been computing the decomposition of the average, but rather the average of the TF of all the trials.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
Shift + click on the cortex surface: Displays the TF decomposition of the selected source.
In the TF figure, it doesn't show as a high value because this file hasn't been normalized.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
Right-click on the brain: Selects the closest vertex and displays the popup menu at the same time. The first three menus are relative to the source that was just clicked.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
Robustness to noise
Same analysis with no noise: Morlet +
Getting rid of the edge effects
On the hard drive
Right click one of the first TF file we computed > File > View file contents.
Structure of the time-frequency files: timefreq_*.mat
TF: [Nsignals x Ntime x Nfreq] matrix containing all the values of the time-frequency decomposition (complex wavelet coefficients, or double values for power/magnitude/Z-score).
TFmask: [Nfreq x Ntime] logical mask indicating the edge effects (0=edge, 1=valid value).
Std: [Nsignals x Ntime x Nfreq] standard deviation if this file is an average.
Comment: String displayed in the database explorer to represent the file.
DataType: From what kind of data this file was computed: {'data', 'results', 'scout, 'matrix'}
Time: [1 x Ntime] Time vector used to estimate this file.
TimeBands: [Ntimebands x 3] Cell array where each line represents a time band:
{'band_name', 'time definition', 'function'}Freqs: For regular frequency binning: vector containing all the frequencies.
If using frequency bands: [Nfreqbands x 3] cell array, where each line represents a frequency band {'band_name', 'frequency definition', 'function'}RefRowNames: Used only for connectivity matrices.
RowNames: [1 x Nsignals] Cell array of strings that describes each row of the TF matrix. In this specific case, it would be the list of all the MEG sensor names. But it could also be a list of names of scouts or clusters.
Measure: Contains the name of the function that was applied right after the computation of the wavelet coefficients. So it represents the type of data contained in the TF matrix. Possible values:
none: No measure applied, TF contains the complex wavelet coefficients.
power: Power for each frequency, ie. the square of the amplitude: abs(coefficients).^2
magnitude: abs(coefficients)
log: 10 * log10( abs(coefficients).^2)
phase: angle(coefficients)
Method: String that identifies the process that generated the file:
{'morlet', 'fft', 'psd', 'hilbert', 'corr', 'cohere', 'granger', 'plv'}DataFile: Initial file from which this file was computed. In the database explorer, the TF file will be shown as a child of this DataFile file.
SurfaceFile / GridLoc / GridAtlas: Source space that was used, only for source files.
nAvg: Number of trials that were averaged to obtain this file.
ColormapType: String, force a specific colormap type to be used when displaying this file.
DisplayUnits: String, force to use specific units when displaying this file.
Options: Options that were selected in the time-frequency options window.
History: List of operations performed on this file (menu File > View file history).
Useful functions
in_bst_timefreq(TimefreqFile): Read a time-frequency file.
in_bst(FileName, TimeWindow): Read any Brainstorm file with the possibility to load only a specific part of the file. "TimeWindow" is an range of time values in seconds: [tStart, tStop].
bst_process('LoadInputFile', FileName, Target, TimeWindow): The most high-level function for reading data files. "Target" is a string with the list of sensor names or types to load.
morlet_transform(): Applies complex Morlet wavelet transform to the time series in input.
References [TODO]
Bertrand O, Tallon-Baudry C (2000)
Oscillatory gamma activity in humans: a possible role for object representation
Int J Psychophysiol, 38(3):211-23Le Van Quyen M, Foucher J, Lachaux J, Rodriguez E, Lutz A, Martinerie J, Varela FJ (2001)
Comparison of Hilbert transform and wavelet methods for the analysis of neuronal synchrony
J Neurosci Methods, 111(2):83-98- Add reference for bandpass+Hilbert: MX Cohen?
Additional documentation
Forum: Time and frequency resolution: http://neuroimage.usc.edu/forums/showthread.php?1848
Delete all your experiments
Before moving to the next tutorial, delete all the time-frequency files you computed in this tutorial. It will make the database structure less confusing for the following tutorials.