|
Size: 7106
Comment:
|
Size: 14043
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = OpenMEEG BEM head model = | = Realistic head model: BEM with OpenMEEG = |
| Line 4: | Line 4: |
| This forward model uses a symmetric boundary element method (symmetric BEM) and was developed by the French public research institute INRIA ([[http://openmeeg.github.io|website]]). It uses three realistic layers (scalp, inner skull, outer skull). The goal of this forward solution is mostly for '''EEG users''', to provide a better model than the existing "3-shell sphere". It is not necessary for MEG users, as the "overlapping spheres" method gives relatively similar results but faster. Nevertheless, for demonstration purpose or in case you want to use the same head model for both EEG and MEG data, we are going to compute this solution on MEG recordings, using the existing TutorialCTF protocol. | This forward model uses a symmetric boundary element method (symmetric BEM) and was developed by the French public research institute INRIA ([[http://openmeeg.github.io|website]]). It uses three realistic layers (scalp, inner skull, outer skull). The goal of this forward solution is mostly for '''EEG users''', to provide more accurate results than the spherical models. It is not necessary for MEG users, as the "overlapping spheres" method gives similar results but much faster. This method is illustrated using the tutorial [[http://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy|EEG and epilepsy]]. |
| Line 6: | Line 6: |
| {{attachment:openmeeg.gif||height="75px",width="274px"}} | {{attachment:openmeeg.gif||width="274px",height="75px"}} |
| Line 14: | Line 14: |
| We need three realistic layers extracted from the subject's MRI (scalp, inner skull, outer skull), plus the source space (cortical surface). Those layers are sometimes complicated to generate. In the case you do not have any way to calculate the inner skull and outer skull surfaces, Brainstorm can generate rough approximations based on the subject's cortex and head surfaces and Colin27's inner and outer skull surfaces.The surfaces created with Brainstorm are by construction non-intersecting. | We need three realistic layers extracted from the subject's MRI (scalp, inner skull, outer skull), plus the source space (cortical surface). These layers are sometimes complicated to generate. In the case you do not have any way to calculate the inner skull and outer skull surfaces, Brainstorm can generate rough approximations based on the subject's cortex and head surfaces and ICBM152's inner and outer skull surfaces. The surfaces created with Brainstorm are by construction non-intersecting. |
| Line 16: | Line 16: |
| 1. Select the TutorialCTF protocol 1. Go to the anatomy view (first button on top of the database explorer) 1. Right-click on Subject01 > Generate BEM surfaces...You can set the approximate number of vertices you want for each layer. For the example, leave the default parameters and click on Ok. 1. Note that those values are very low and could lead to incoherent results. However, using more vertices on each layer can increase a lot the amount of memory necessary and the computation time. If you can afford it computationally, you should use a minimum of '''2000 vertices''' per layer. Note that you may not get the number of vertices you are asking for, the algorithm generating those surfaces uses a fixed set of apolar spheres with the following number of vertices: 273, 362, 482, 642, 812, 1082, 1442, 1922, 2432, 2562, 3242, 4322...<<BR>><<BR>> {{attachment:bemPopup.gif}} {{attachment:bemOptions.gif}} 1. At the end of the process, three new surfaces are added to the database:<<BR>><<BR>> {{attachment:bemFiles.gif}} {{attachment:bem3d.gif}} |
1. Select the protocol TutorialEpilepsy. 1. Go to the anatomy view (first button on top of the database explorer). 1. Right-click on the subject folder > '''Generate BEM surfaces'''. You can set the approximate number of vertices you want for each layer. For this example, leave the default parameters and click on Ok. <<BR>><<BR>> {{attachment:bemPopup.gif}} {{attachment:bemOptions.gif}} 1. At the end of the process, three new surfaces are added to the database:<<BR>><<BR>> {{attachment:bemFiles.gif}} {{attachment:bem3d.gif||width="299",height="260"}} ==== How to set the number of vertices per layer ==== * The more vertices for each layer, the more accurate your head model. If there are not enough vertices, OpenMEEG may consider that the surfaces are intersecting and crash before the computation, or it may finish but at the end the model may be unstable with a few vertices with values much higher than all the others. * On the other hand, using more vertices on each layer increases a lot the amount of memory necessary and the computation time. If your system hangs, if it takes 24hrs to run or if you get "Out of memory" errors, you should decrease the number of vertices for each layer. * The default values are high (1922 vertices per layer), to ensure that by default the results are good. If you can afford it computationally, you should use these '''1922 vertices''' per layer. If you have to decrease these values, you can try for instance with '''1082 vertices '''(scalp) and '''642 vertices '''(outer skull and innser skull). * If you are using the default ICBM152 anatomy, you do not need to run this, you can directly use the head/outerskull/innerskull surfaces in the the default anatomy folder. The number of vertices is much lower, but we know this anatomy to be stable, you should not need to recalculate new surfaces with more vertices. |
| Line 23: | Line 28: |
| 1. Switch to the view "Functional data (sorted by subjects)" 1. Right-click on Subject01 / Right > Compute head model. Select OpenMEEG BEM in the list. Click on Run.<<BR>><<BR>> {{attachment:headmodeler.gif}} {{attachment:openmeegOptions.gif}} |
1. Switch to the view "Functional data (sorted by subjects)", 2nd button above the database explore. 1. Right-click on the channel file > '''Compute head model'''. Select '''OpenMEEG BEM''' in the list. <<BR>><<BR>> {{attachment:headmodeler.gif||width="670",height="326"}} |
| Line 26: | Line 31: |
| * '''BEM Layers & conductivities''': In this panel you can select how many layers you want to use for the BEM model. The possible options in our case are Scalp (=head), Skull (=outer skull), and Brain (=inner skull). The values on the right are the relative conductivities of each layer. | * '''BEM Layers & conductivities''': You can select how many layers you want to use for the BEM model. The possible options in our case are Scalp (=head), Skull (=outer skull), and Brain (=inner skull). The values on the right are the relative conductivities of each layer. |
| Line 30: | Line 35: |
| * '''Use adaptive integration''': This produces a much more accurate solution, mostly in the cases of dipoles (vertices of the cortex surface) that are very close to the inner skull layer. You should keep this option selected. * '''Process dipoles by blocks''': If you consistently run into crashes, it's probably because you don't have enough memory available. You can try to process the dipoles by smaller blocks. It would be longer but would probably work. Do not select unless you cannot compute the solution with having memory errors. 1. Download OpenMEEG software? Yes.<<BR>> This will download the latest version of OpenMEEG binaries for your operating system from INRIA website, and install it in Brainstorm user folder (~username/.brainstorm/openmeeg/)<<BR>><<BR>> {{attachment:openmeegDownload.gif}} 1. Once downloaded, the calculation of the head model will start automatically. You may wait for a very long time. Be patient, it's worth it...<<BR>><<BR>> {{attachment:openmeegWait.gif}} 1. You have now a new head model in your condition, that you can use exactly the same way you would use your previous "Overlapping sphere" model.<<BR>><<BR>> {{attachment:treeFinal.gif}} |
* '''Use adaptive integration''': This produces a more accurate solution, mostly in the cases of dipoles (vertices of the cortex surface) that are very close to the inner skull layer. Keep this option selected. * '''Process dipoles by blocks''': If you consistently run into crashes, it could be because you don't have enough memory available. You can try to process the dipoles by smaller blocks. It would be longer but could work. Select this option only if you have memory errors. 1. Download OpenMEEG software? '''Yes'''.<<BR>> This will download the latest version of the OpenMEEG binaries for your operating system from INRIA website, and install it in Brainstorm user folder (~username/.brainstorm/openmeeg/)<<BR>><<BR>> {{attachment:openmeegDownload.gif}} 1. Then the calculation of the head model will automatically start. You may wait for a very long time. Be patient, it's worth it...<<BR>><<BR>> {{attachment:openmeegWait.gif}} 1. You have now a new head model for this subject.<<BR>><<BR>> {{attachment:treeFinal.gif}} |
| Line 36: | Line 41: |
| == Additional discussions on the forum == Download problems: |
== Warning: Dipoles outside == OpenMEEG BEM solver may be unstable when the dipoles (= the vertices of the cortex surface) are outside or too close to the innermost layer (= the brain/inner skull surface). Brainstorm tries to detect for which dipoles OpenMEEG could return wrong values, and displays a warning if any: |
| Line 39: | Line 44: |
| * [[http://neuroimage.usc.edu/forums/showthread.php?1066-Impossible-to-download-OpenMEEG|http://neuroimage.usc.edu/forums/showthread.php?1066]] | {{attachment:warning_dipout.gif}} |
| Line 41: | Line 46: |
| Binary issues / Error #-1073741515: | This doesn't mean that these points would necessarily cause problems in the BEM forward model, but it is important to be aware that they might, and check this carefully. First, evaluate visually the '''intersections between the cortex and the innerskull''' surfaces: display both in a 3D view by double-clicking on them, then from the Surface tab, set a bright color for the cortex and 0% transparency for the innerskull. This image may reveal that some parts of the cortex are sticking out of the inner skull surface. |
| Line 43: | Line 48: |
| * [[http://neuroimage.usc.edu/forums/showthread.php?744-Problem-with-BST-and-Windows-7|http://neuroimage.usc.edu/forums/showthread.php?744]] * http://neuroimage.usc.edu/forums/showthread.php?767 * http://neuroimage.usc.edu/forums/showthread.php?1145 |
{{attachment:warning_dipout2.png}} |
| Line 47: | Line 50: |
| Library issues / Error #1: | To fix the surfaces, two solutions: |
| Line 49: | Line 52: |
| * http://neuroimage.usc.edu/forums/showthread.php?1309-compute-head-model-issue | * '''Fix the inner skull''' surface by '''recomputing the BEM surfaces with more vertices'''. With more points, the envelope would have more details to wrap better the cortex surface. Always try this option first. The figures below show 12 dipoles outside the 482V inner skull, while all the dipoles fit in the 1922V inner skull.<<BR>><<BR>> {{attachment:warning_dipout4.png}} * '''Fix the cortex''' surface with the popup menu '''Force inside skull''': pushes the vertices that are outside of the inner skull surface into it. This can be a solution to quickly get rid of the surface intersection and avoid mathematical issues in the computation of the BEM solution. However, this is only adapted to correct for minor topological issues, when the cortex surface is almost tangential to the skull and only a few cortex vertices appear outside. When selecting this option, the original surface is displayed with red dots for the edited vertices, together with the new fixed surface. <<BR>><<BR>> {{attachment:warning_dipout3.png}} |
| Line 51: | Line 55: |
| Memory allocation failed: | Note that this is only a '''WARNING''': even with many suspicious points, the BEM forward model might be entirely valid. You may decide to keep the surfaces unmodified, and go back to fixing them only if you observe abnormal lead fields or source values at these locations. |
| Line 53: | Line 57: |
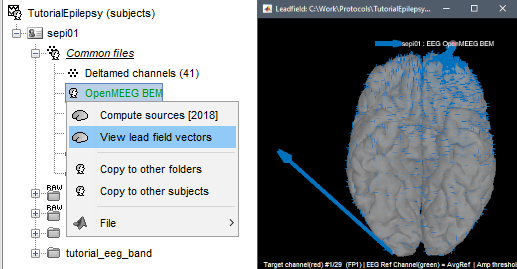
| * http://neuroimage.usc.edu/forums/showthread.php?1138 | To display the forward fields, right-click on the BEM head model > '''View lead field vectors'''. Abnormal displays may look like the one below: one or a few arrows are much longer than all the others. You may need to adjust the relative size of the lead field arrows in order to see something - press H for help. In this case, one of the occipital dipoles that was initially detected as a possible problem shows an abnormality that may impact your source analysis: the surfaces should be fixed and the forward model recomputed. |
| Line 55: | Line 59: |
| Intersecting meshes: | {{attachment:warning_dipout5.png}} |
| Line 57: | Line 61: |
| * http://neuroimage.usc.edu/forums/showthread.php?1030 * http://neuroimage.usc.edu/forums/showthread.php?1305 |
The example used to illustrated this section is the [[https://neuroimage.usc.edu/brainstorm/Tutorials/Epilepsy|EEG/Epilepsy tutorial]], using BEM surfaces with 642/482/482 vertices. Everything can be reproduced easily. |
| Line 60: | Line 63: |
| Format of the Gain matrix in the headmodel file: | == Errors == OpenMEEG may crash for various reasons: we tried to list the possible causes here. |
| Line 62: | Line 66: |
| * http://neuroimage.usc.edu/forums/showthread.php?918 * http://neuroimage.usc.edu/forums/showthread.php?1353 |
=== Linux === * OpenMEEG error #1:<<BR>>./om_assemble: /usr/local/MATLAB/R2013b/sys/os/glnxa64/libstdc++.so.6: version `GLIBCXX_3.4.19’ not found (required by ./om_assemble) * '''Cause''': Your version of Matlab uses a version of library libstdc++ that is older than the one required by OpenMEEG. * '''Solution''': If your operating system is up-to-date, you should have a newer version available on your hard drive. You need to tell Matlab to use the local version of libstdc++ instead of the one shipped with Matlab. In order to do this, open a terminal and start Matlab in the following way: <<BR>>LD_PRELOAD=/usr/lib/x86_64-linux-gnu/libstdc++.so.6 /usr/local/bin/matlab * You might need to adjust the path to the Matlab executable (get it by typing "which matlab") or the libstdc++ library (type "locate libstdc++.so.6" and pick the one in /usr/lib) * Library issues: https://neuroimage.usc.edu/forums/t/openmeeg-headmodel-error/7214/18 === MacOS === * You get a system error: your computer doesn't want to execute om_assemble or any other om_* program because the '''developers are unknown'''. * '''Solution''': Go to the OpenMEEG installation folder (most likely somewhere in $HOME/.brainstorm) and explicitly tell your system that each one of these om_* executables are OK to execute (look it up if you don't find an obvious way to do so). === Windows === * OpenMEEG error #-1073741515: * '''Cause''': Missing package in Windows. * '''Solution''': Install the "Microsoft Visual C++ 2010 Redistributable Package": http://www.microsoft.com/download/en/details.aspx?id=14632 * OpenMEEG crashes on a 32bit operating system: * '''Cause''': OpenMEEG needs to address a lot of memory, which is not be possible on a 32bit OS. * '''Solution''': Use a different computer. === All systems === * OpenMEEG cannot be downloaded: * '''Cause''': You have a proxy that is not configured correctly with Matlab. * '''Solution''': Download OpenMEEG manually (http://openmeeg.gforge.inria.fr/download/) and use the Brainstorm menu Update > Update OpenMEEG > Install. * The default version of OpenMEEG is not supported on your system: * '''Solution''': Download another OpenMEEG release manually (http://openmeeg.gforge.inria.fr/download/) and use the Brainstorm menu Update > Update OpenMEEG > Install. * OpenMEEG crashes after a long computation time or runs forever: * '''Cause''': Your system possibly ran out of memory. Open a resource monitor on your computer, start again the forward model computation and check the evolution of the memory usage. If it goes above 90-95%, you are reaching the maximum capacity of your system. It might start swapping (using the hard drive instead of the RAM memory, which is awfully slow) or simply crash. * '''Solution''': Use a simplified model: BEM layers with less vertices, and/or less dipoles in your source space (downsample the cortex surface), and/or different OpenMEEG options. * Intersecting meshes or triangles: * '''Cause''': OpenMEEG runs only on closed, regular, very smooth surfaces and non-intersecting surfaces. A surface can be self-intersecting or two layers can be crossing each other even if you cannot see it when displaying them with Brainstorm. * '''Solution''':Iif you were trying to use BEM surfaces from a different source, try computing them from Brainstorm using the menu "Generate BEM surfaces". If it crashes with BEM surfaces generated with Brainstorm, try increasing the number of vertices on the various layers, or try different combination of values for the various layers. === Solution not found === If you cannot find a solution, please post a bug report on the Brainstorm [[https://neuroimage.usc.edu/forums/|user forum]]: * Copy-paste the full error message from the Matlab command window. * Zip the contents of the folder ''$HOME/.brainstorm/tmp'' immediately after the crash, upload it somewhere and include the download link in your bug report. * Include a screen capture of a 3D figure showing the sensors and all the BEM surfaces. == Additional documentation == Tutorial "Head modeling": https://neuroimage.usc.edu/brainstorm/Tutorials/HeadModel |
| Line 70: | Line 113: |
| * Gramfort, T. Papadopoulo, E. Olivi, M. Clerc. OpenMEEG: opensource software for quasistatic bioelectromagnetics, [[http://www.biomedical-engineering-online.com/content/9/1/45|BioMedical Engineering OnLine 45:9, 2010]] * Kybic J, Clerc M, Abboud T, Faugeras O, Keriven R, Papadopoulo T. A common formalism for the integral formulations of the forward EEG problem. [[http://ieeexplore.ieee.org/xpls/abs_all.jsp?isnumber=30034&arnumber=1375158&count=10&index=1|IEEE Transactions on Medical Imaging, 24:12-28, 2005.]] [[ftp://ftp-sop.inria.fr/odyssee/Publications/2005/kybic-clerc-etal:05.pdf|[pdf]]] |
* Gramfort A, Papadopoulo T, Olivi E, Clerc M<<BR>>[[http://www.biomedical-engineering-online.com/content/9/1/45|OpenMEEG: opensource software for quasistatic bioelectromagnetics]]<<BR>>BioMedical Engineering OnLine 45:9, 2010 * Kybic J, Clerc M, Abboud T, Faugeras O, Keriven R, Papadopoulo T<<BR>>[[http://ieeexplore.ieee.org/xpls/abs_all.jsp?isnumber=30034&arnumber=1375158&count=10&index=1|A common formalism for the integral formulations of the forward EEG problem]]<<BR>>IEEE Transactions on Medical Imaging, 24:12-28, 2005 | [[ftp://ftp-sop.inria.fr/odyssee/Publications/2005/kybic-clerc-etal:05.pdf|pdf]] |
| Line 73: | Line 116: |
| == Feedback == <<EmbedContent(http://neuroimage.usc.edu/brainstorm3_register/get_feedback.php?Tutorials/TutBem)>> |
<<EmbedContent(http://neuroimage.usc.edu/bst/get_feedback.php?Tutorials/TutBem)>> |
Realistic head model: BEM with OpenMEEG
Authors: Francois Tadel, Alexandre Gramfort
This forward model uses a symmetric boundary element method (symmetric BEM) and was developed by the French public research institute INRIA (website). It uses three realistic layers (scalp, inner skull, outer skull). The goal of this forward solution is mostly for EEG users, to provide more accurate results than the spherical models. It is not necessary for MEG users, as the "overlapping spheres" method gives similar results but much faster. This method is illustrated using the tutorial EEG and epilepsy.

Contents
Requirements
On Windows 64bit systems, you need to install an extra Microsoft software package to run OpenMEEG: Microsoft Visual C++ 2010 Redistributable Package (x64).
http://www.microsoft.com/en-us/download/details.aspx?id=14632
BEM surfaces
We need three realistic layers extracted from the subject's MRI (scalp, inner skull, outer skull), plus the source space (cortical surface). These layers are sometimes complicated to generate. In the case you do not have any way to calculate the inner skull and outer skull surfaces, Brainstorm can generate rough approximations based on the subject's cortex and head surfaces and ICBM152's inner and outer skull surfaces. The surfaces created with Brainstorm are by construction non-intersecting.
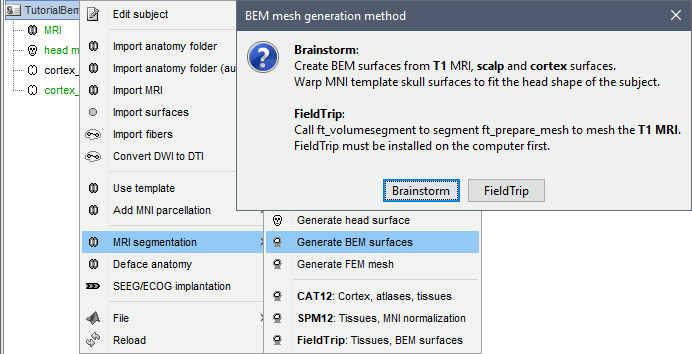
Select the protocol TutorialEpilepsy.
- Go to the anatomy view (first button on top of the database explorer).
Right-click on the subject folder > Generate BEM surfaces. You can set the approximate number of vertices you want for each layer. For this example, leave the default parameters and click on Ok.
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
At the end of the process, three new surfaces are added to the database:
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
![[ATTACH] [ATTACH]](/moin_static1911/brainstorm1/img/attach.png)
How to set the number of vertices per layer
- The more vertices for each layer, the more accurate your head model. If there are not enough vertices, OpenMEEG may consider that the surfaces are intersecting and crash before the computation, or it may finish but at the end the model may be unstable with a few vertices with values much higher than all the others.
- On the other hand, using more vertices on each layer increases a lot the amount of memory necessary and the computation time. If your system hangs, if it takes 24hrs to run or if you get "Out of memory" errors, you should decrease the number of vertices for each layer.
The default values are high (1922 vertices per layer), to ensure that by default the results are good. If you can afford it computationally, you should use these 1922 vertices per layer. If you have to decrease these values, you can try for instance with 1082 vertices (scalp) and 642 vertices (outer skull and innser skull).
- If you are using the default ICBM152 anatomy, you do not need to run this, you can directly use the head/outerskull/innerskull surfaces in the the default anatomy folder. The number of vertices is much lower, but we know this anatomy to be stable, you should not need to recalculate new surfaces with more vertices.
Forward model
- Switch to the view "Functional data (sorted by subjects)", 2nd button above the database explore.
Right-click on the channel file > Compute head model. Select OpenMEEG BEM in the list.
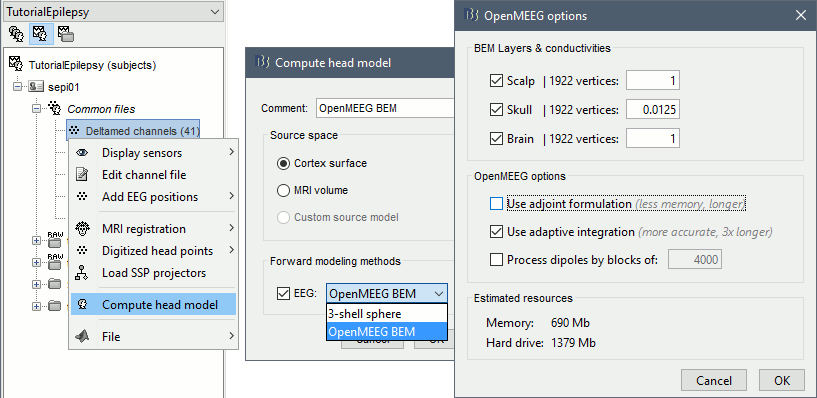
- Explanations of the options:
BEM Layers & conductivities: You can select how many layers you want to use for the BEM model. The possible options in our case are Scalp (=head), Skull (=outer skull), and Brain (=inner skull). The values on the right are the relative conductivities of each layer.
- For EEG: Use all the layers
- For MEG: Use only the inner skull layer
Use adjoint formulation: Decrease significantly the amount of memory that is necessary in memory and on the hard drive, but much longer.
Use adaptive integration: This produces a more accurate solution, mostly in the cases of dipoles (vertices of the cortex surface) that are very close to the inner skull layer. Keep this option selected.
Process dipoles by blocks: If you consistently run into crashes, it could be because you don't have enough memory available. You can try to process the dipoles by smaller blocks. It would be longer but could work. Select this option only if you have memory errors.
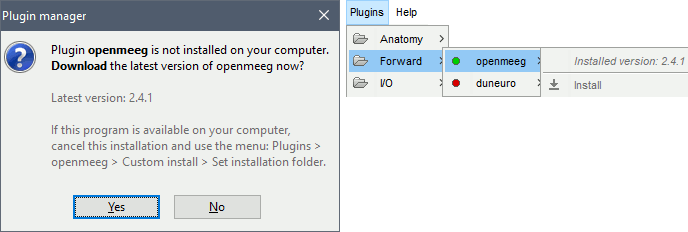
Download OpenMEEG software? Yes.
This will download the latest version of the OpenMEEG binaries for your operating system from INRIA website, and install it in Brainstorm user folder (~username/.brainstorm/openmeeg/)
Then the calculation of the head model will automatically start. You may wait for a very long time. Be patient, it's worth it...

You have now a new head model for this subject.
Warning: Dipoles outside
OpenMEEG BEM solver may be unstable when the dipoles (= the vertices of the cortex surface) are outside or too close to the innermost layer (= the brain/inner skull surface). Brainstorm tries to detect for which dipoles OpenMEEG could return wrong values, and displays a warning if any:
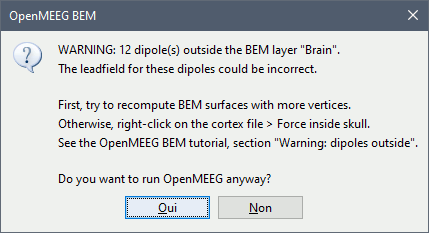
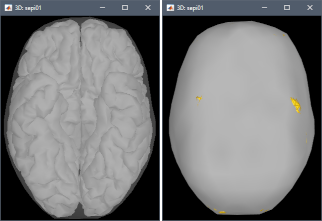
This doesn't mean that these points would necessarily cause problems in the BEM forward model, but it is important to be aware that they might, and check this carefully. First, evaluate visually the intersections between the cortex and the innerskull surfaces: display both in a 3D view by double-clicking on them, then from the Surface tab, set a bright color for the cortex and 0% transparency for the innerskull. This image may reveal that some parts of the cortex are sticking out of the inner skull surface.
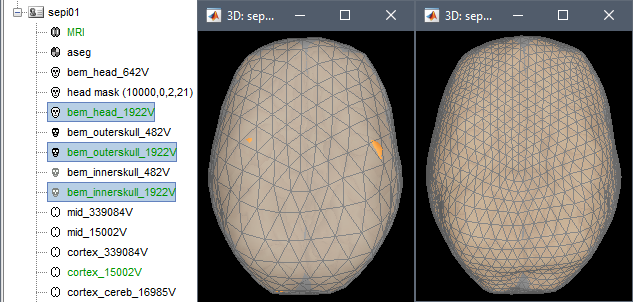
To fix the surfaces, two solutions:
Fix the inner skull surface by recomputing the BEM surfaces with more vertices. With more points, the envelope would have more details to wrap better the cortex surface. Always try this option first. The figures below show 12 dipoles outside the 482V inner skull, while all the dipoles fit in the 1922V inner skull.
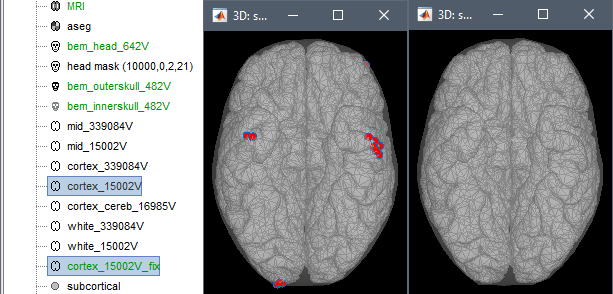
Fix the cortex surface with the popup menu Force inside skull: pushes the vertices that are outside of the inner skull surface into it. This can be a solution to quickly get rid of the surface intersection and avoid mathematical issues in the computation of the BEM solution. However, this is only adapted to correct for minor topological issues, when the cortex surface is almost tangential to the skull and only a few cortex vertices appear outside. When selecting this option, the original surface is displayed with red dots for the edited vertices, together with the new fixed surface.
Note that this is only a WARNING: even with many suspicious points, the BEM forward model might be entirely valid. You may decide to keep the surfaces unmodified, and go back to fixing them only if you observe abnormal lead fields or source values at these locations.
To display the forward fields, right-click on the BEM head model > View lead field vectors. Abnormal displays may look like the one below: one or a few arrows are much longer than all the others. You may need to adjust the relative size of the lead field arrows in order to see something - press H for help. In this case, one of the occipital dipoles that was initially detected as a possible problem shows an abnormality that may impact your source analysis: the surfaces should be fixed and the forward model recomputed.
The example used to illustrated this section is the EEG/Epilepsy tutorial, using BEM surfaces with 642/482/482 vertices. Everything can be reproduced easily.
Errors
OpenMEEG may crash for various reasons: we tried to list the possible causes here.
Linux
OpenMEEG error #1:
./om_assemble: /usr/local/MATLAB/R2013b/sys/os/glnxa64/libstdc++.so.6: version `GLIBCXX_3.4.19’ not found (required by ./om_assemble)Cause: Your version of Matlab uses a version of library libstdc++ that is older than the one required by OpenMEEG.
Solution: If your operating system is up-to-date, you should have a newer version available on your hard drive. You need to tell Matlab to use the local version of libstdc++ instead of the one shipped with Matlab. In order to do this, open a terminal and start Matlab in the following way:
LD_PRELOAD=/usr/lib/x86_64-linux-gnu/libstdc++.so.6 /usr/local/bin/matlab- You might need to adjust the path to the Matlab executable (get it by typing "which matlab") or the libstdc++ library (type "locate libstdc++.so.6" and pick the one in /usr/lib)
Library issues: https://neuroimage.usc.edu/forums/t/openmeeg-headmodel-error/7214/18
MacOS
You get a system error: your computer doesn't want to execute om_assemble or any other om_* program because the developers are unknown.
Solution: Go to the OpenMEEG installation folder (most likely somewhere in $HOME/.brainstorm) and explicitly tell your system that each one of these om_* executables are OK to execute (look it up if you don't find an obvious way to do so).
Windows
- OpenMEEG error #-1073741515:
Cause: Missing package in Windows.
Solution: Install the "Microsoft Visual C++ 2010 Redistributable Package": http://www.microsoft.com/download/en/details.aspx?id=14632
- OpenMEEG crashes on a 32bit operating system:
Cause: OpenMEEG needs to address a lot of memory, which is not be possible on a 32bit OS.
Solution: Use a different computer.
All systems
- OpenMEEG cannot be downloaded:
Cause: You have a proxy that is not configured correctly with Matlab.
Solution: Download OpenMEEG manually (http://openmeeg.gforge.inria.fr/download/) and use the Brainstorm menu Update > Update OpenMEEG > Install.
- The default version of OpenMEEG is not supported on your system:
Solution: Download another OpenMEEG release manually (http://openmeeg.gforge.inria.fr/download/) and use the Brainstorm menu Update > Update OpenMEEG > Install.
- OpenMEEG crashes after a long computation time or runs forever:
Cause: Your system possibly ran out of memory. Open a resource monitor on your computer, start again the forward model computation and check the evolution of the memory usage. If it goes above 90-95%, you are reaching the maximum capacity of your system. It might start swapping (using the hard drive instead of the RAM memory, which is awfully slow) or simply crash.
Solution: Use a simplified model: BEM layers with less vertices, and/or less dipoles in your source space (downsample the cortex surface), and/or different OpenMEEG options.
- Intersecting meshes or triangles:
Cause: OpenMEEG runs only on closed, regular, very smooth surfaces and non-intersecting surfaces. A surface can be self-intersecting or two layers can be crossing each other even if you cannot see it when displaying them with Brainstorm.
Solution:Iif you were trying to use BEM surfaces from a different source, try computing them from Brainstorm using the menu "Generate BEM surfaces". If it crashes with BEM surfaces generated with Brainstorm, try increasing the number of vertices on the various layers, or try different combination of values for the various layers.
Solution not found
If you cannot find a solution, please post a bug report on the Brainstorm user forum:
- Copy-paste the full error message from the Matlab command window.
Zip the contents of the folder $HOME/.brainstorm/tmp immediately after the crash, upload it somewhere and include the download link in your bug report.
- Include a screen capture of a 3D figure showing the sensors and all the BEM surfaces.
Additional documentation
Tutorial "Head modeling": https://neuroimage.usc.edu/brainstorm/Tutorials/HeadModel
License
OpenMEEG is distributed under the French opensource license CeCILL-B. It is intended to give users the freedom to modify and redistribute the software. It is therefore compatible with popular opensource licenses such as the GPL and BSD licenses. The CeCILL-B license imposes to anybody distributing a software incorporating OpenMEEG the obligation to give credits (by citing the appropriate publications), in order for all contributions to be properly identified and acknowledged.
The references to be acknowledged are:
Gramfort A, Papadopoulo T, Olivi E, Clerc M
OpenMEEG: opensource software for quasistatic bioelectromagnetics
BioMedical Engineering OnLine 45:9, 2010Kybic J, Clerc M, Abboud T, Faugeras O, Keriven R, Papadopoulo T
A common formalism for the integral formulations of the forward EEG problem
IEEE Transactions on Medical Imaging, 24:12-28, 2005 | pdf
