|
Size: 1169
Comment:
|
Size: 34824
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 1: | Line 1: |
| = Tutorial: Explore the recordings = This tutorial is based on the ''TutorialCTF'' protocol, and will show you how to explore efficiently the sensory response to an electric stimulation of the fingers. The two previous tutorials explain how to create this tutorial and describe the data that are used in it. <<TableOfContents(2)>> == Display modes == Several display modes are available for recordings. They are all accessible with a right-click.The first three menus (MEG, Stimulation input, Video Time), represent all the different types of channels that were found in the channel file. You can check that with Channel Edit (right-click on channel file > Edit). |
'''[WARNING: This page is part of the old tutorials, please refer to the [[Tutorials|new documentation]]]'''<<BR>><<BR>><<BR>> = Tutorial 5: Exploring the recordings = ''Authors: Francois Tadel, John C Mosher, Sylvain Baillet'' This tutorial is based on the ''TutorialCTF'' protocol, and will show you how to explore efficiently the sensory response to an electric stimulation of the fingers. <<TableOfContents(2,2)>> == Display recordings == Several display modes are available for recordings. They are all accessible with a right-click.The first four menus (MEG, MEG REF, Stim, Video), represent all the different types of channels that were found in the channel file. You can check that with Channel Edit (right-click on channel file > Edit). We are only interested in the "MEG" in this tutorial. Select Left / ERF > MEG > Display time series. |
| Line 11: | Line 15: |
| <<BR>>Now repeat this operation several times to display all the available modes: ''Display time series, 3D sensor cap, 2D sensor cap, 2D disc'' and ''2D Layout''. You should now see all the following figures. {{attachment:dataTimeSeries.gif}} {{attachment:data2Dlayout.gif}} {{attachment:data3Dcap.gif}} {{attachment:data2Dcap.gif}} {{attachment:data2Ddisc.gif}} |
<<BR>>Now repeat this operation several times to display all the available modes: ''Display time series, 3D sensor cap, 2D sensor cap, 2D disc'' and ''2D Layout''. You should now see all the figures displayed below. Keep them opened for the next few paragraphs, you will learn how to manipulate multiple views of the same data. === Display time series === Amplitude fluctuations over time for all the sensors. {{attachment:dataTimeSeries.gif}} Buttons available on the figure: * '''[<]''' and '''[>]''': Horizontal zoom around the current time point * '''[^]''' and '''[v]''': Vertical zoom * '''[Flip +/-'''''']''': Exchange the direction of the Y axis, useful in clinical EEG * '''[...'''''']''': Set the amplitude range manually, ie. the limits of the Y axis * '''[AS'''''']''': Amplitude auto-scale. When selected, the amplitude range (Y axis limits) is automatically updated when the figure is updated. This is useful mostly when reviewing continuous files or individual trials, it will be illustrated in other tutorials. List of mouse and keyboard operations for this type of figure (most of the operations are detailed later): * '''Mouse wheel''': Horizontal zoom * '''CTRL+mouse wheel''': Vertical zoom * '''Right click + move''': Move in zoomed figure * '''Left click + move''': Time selection * '''Left-click''': Set current time * '''Left-click on a line''': Select the corresponding sensor * ''' Shift + Left-click''': Force setting the current time, even when clicking on a sensor * '''Right-click''': Popup menu * '''Double-click''': Restore initial view (ie. unzoom) * '''Keyboard arrows''': Change current time * '''PageUp / PageDown''': Change current time (10 samples at a time) * __Notes for Mac users__: * All the actions involving the mouse wheel can be performed on MacBook laptops by moving up and down two fingers on the pad. * "PageUp" or "PageDown" keys are obtained by pressing the "Fn" key together with the arrows UP or DOWN === 3D Sensor cap / 2D Sensor cap / 2D Disc === The recordings at a given time instant and interpolated over a 2D or 3D surface. {{attachment:data3Dcap.gif||height="175px",width="204px"}} {{attachment:data2Dcap.gif||height="174px",width="203px"}} {{attachment:data2Ddisc.gif||height="173px",width="201px"}} Keyboard and mouse shortcuts: * '''Mouse wheel''': Zoom / unzoom * '''Left click + move''': Rotate (3D only) * '''Middle click + move''': Or left+right click + move: Move in zoomed figure * '''Right click + move''': Select sensors (2D only, when sensors are visible, see below) * '''Right click''': Popup menu * '''Left click on the colorbar + move''': Change contrast (up/down) and brightness (left/right) * '''Control + E''': Display the channels markers and/or names * '''Keyboard arrows''': Change current time * '''PageUp / PageDown''': Change current time (10 samples at a time) / Mac: Fn+UP/Fn+Down === 2D layout === The time course of each channel is drawn at the actual position of the electrode, projected in 2D the same way as ''2D Sensor cap''. The light gray lines represent the zero amplitude (horizontal) and the current time (vertical lines). {{attachment:data2Dlayout.gif}} {{attachment:data2DlayoutWhite.gif}} {{attachment:data2DlayoutMenu.gif}} Only a part of the full time window is displayed for each channel, before and after the current time. The length of this time window can be modified either with the mouse shortcut Control+wheel, or with the 2D Layout options, in the figure popup menu. Other options are also available in the popup menu. * '''Mouse wheel''': Zoom / unzoom * '''Middle click + move''': (or left+right click + move) Move into the zoomed figure * '''Shift + mouse wheel''': Increase/decrease the amplitude gain of the channels * '''Control + mouse wheel''': Increase/decrease the length of the time window around the current time * '''Click on a line''': Select a sensor * '''Right click + move''': Select a group of sensors * '''Shift + click on a line''': Select one sensor and unselect all the others<<BR>>Display the time-frequency decomposition for the selected sensor, when available * '''Right-click''': Display popup menu. <<BR>>Sub-menu "2DLayout options" offer some configuration options for this type of figure. * '''Control + E''': Display/hide the channels names * '''Keyboard arrows''': Change current time * '''PageUp / PageDown''': Change current time (10 samples at a time) / Mac: Fn+UP/Fn+Down === Magnetic interpolation === Some of the views (3D Sensor cap / 2D Sensor cap / 2D Disc), are by default re-interpolating the field that is recorded by the sensors to get much smoother displays. A simple inverse problem + forward problem are solved to reconstruct the magnetic fields on a high-resolution surface. The menu '''"No magnetic interpolation"''' offer the same views, but without using this reconstruction of the magnetic field, and performing instead a spatial interpolation of the values between the sensors. {{attachment:data3DcapRaw.gif|data3Dcap.gif|height="175px",width="204px"}} {{attachment:data2DcapRaw.gif|data2Dcap.gif|height="174px",width="203px"}} {{attachment:data2DdiscRaw.gif|data2Ddisc.gif|height="173px",width="201px"}} == Graphic bugs == If you observe any graphic problem with these displays, there might be an issue with the OpenGL drivers. You may try the following options: * Update the drivers for your graphics card. * Upgrade your version of Matlab. * Run the compiled version of Brainstorm (see [[http://neuroimage.usc.edu/brainstorm/Installation|Installation]]). * Turn off the OpenGL hardware acceleration: Menu File > Edit preferences > ''Software'' or ''Disabled''. * Send a bug report to the Mathworks. |
| Line 18: | Line 130: |
| The time window and the current time instant are centralized and managed from the top of the Brainstorm window. You can use the text box or the buttons to change the current time value. All the figures always stay synchronized, you cannot display the 2D disc topography at t=10ms and the 3D sensor cap topography at t=50ms at the same time. It might look too restrictive at the beginning, but the contrary would have made the interface much too complex; we wanted to keep it as simple and intuitive as possible. {{attachment:timeWindow.gif}} The time window panel is not the best way to explore your recordings. There are many other shortcuts that are much faster to navigate through the time. * '''Click on ''''''time series'''''' figure''':<<BR>>Left-click in any white area of the figure: it will move the time cursor at this point and update all the other figures to match this new value. Don't click on a black line, it would select the corresponding channel instead of moving the time cursor. * '''Press the keyboard arrows''': <<BR>>Click on any data figure and try to press several time the left and right arrows, and also the ''PageUp ''and ''PageDown ''keys. This feature is really useful when you want to quickly see how the recordings are changing in time. Try holding the arrow keys for a while. It might be a bit slow because you have many figures to refresh at each time change. It is much faster if you have only one 2D figure to refresh. '''To remember''': An efficient and quick way to review your recordings just after importation is to: 1. Open a Time series window (double click on recordings file) 1. Click on the time of interest (eg. first response peak) 1. Open a topography view of your choice (2D disc, 2D sensor cap, or 3D sensor cap)<<BR>>You can do this by right-clicking on the time series figure > MEG Topography (or Ctrl+T keyboard shortcut). 1. Use the left/right arrows, and PageUp/PageDown keys (or Fn+Up / Fn+Down on a Mac) |
|
| Line 19: | Line 149: |
| You can select some channels, and then display them separately or mark them as good or bad. 1. Now close all the figures except the ''Time series'' and the ''2D Sensor cap'' topography (if you don't remember which one it was, close everything and open again these two figures). 1. On ''2D Sensor cap'' figure: right-click anywhere on the window ''> Channels > Display sensors''. You'll see white dots representing the center of each MEG coil. Note that there is keyboard shortcut indicated for this menu: ''Ctrl + E''. 1. Press'' Ctrl + E'' several times and see what happens. Come back to the display with only the sensors markers (no labels). 1. Click on some white dots. * They turn red and the corresponding lines in the time series window also turn red * Left-click on the lines in the time series window: it also selects the sensors * Right-click on a red line in time series figure: shows the name and index of the selected channel * Click on a selected (red) point or line: the sensor is deselected. * To deselect all the sensors, press ''Escape'' or ''right-click > Channels > Reset selection'' 1. Now select randomly three sensors.<<BR>><<BR>> {{attachment:channelSelection.gif||height="174px",width="545px"}} 1. Right-click > Channels > View selected (Shortcut = Enter key). A new window is created, with only these three sensors, whose names are indicated in a legend box (you can move this legend if needed).<<BR>><<BR>> {{attachment:channelPopupMenu.gif||height="143px",width="327px"}} {{attachment:channelViewSelected.gif||height="121px",width="295px"}} 1. Close that last figure. In the 2D topography figure, right-click and move the mouse to select a group of sensors, just to remember that this feature exists.<<BR>><<BR>> {{attachment:channelGroupSelection.gif}} == Time series in columns == There are two display modes for the MEG signals: "butterfly", what we've seen until now, or "column". Double click on Left/ERP to open a time series figure. To switch from the current mode to the "column" mode, click on {{attachment:buttonTsCol.gif}} in the Record tab. * The figure you get is unreadable because it contains the 151 sensors in the same figure. There are several ways to get a readable display: you need to select a subset of the channels, zoom and/or to increase the gain of the channels.<<BR>><<BR>> {{attachment:tsColumn.gif}} * Start by maximizing the size of the figure. If you have a large screen, you might be already able to see something. * '''Channel gain''': You can increase/decrease the amplitude of the traces in different ways: * Click on the up/down buttons on the right side of the figure * Click on the "..." button on the right of the window to set manually the gain value for each vertical unit in the graph (distance between two horizontal lines). * Press the "+" or "-" keys on your keyboard * Hold the SHIFT key down, and use the mouse wheel to zoom/unzoom * '''Vertical zoom''': Hold the CTRL key down, and use the mouse wheel to zoom in or out. * '''Horizontal zoom''': * Click on the "<" and ">" buttons at the bottom of the window * Use the mouse wheel to zoom in or out. * '''Move in the zoomed figure''': Hold down left and right buttons of the mouse, and move the mouse. * You can reach some display results that are already a little more interesting:<<BR>><<BR>> {{attachment:tsColumnZoom.gif}} * '''Reset initial view''': Double-click == Montages == A better way to review a large number of MEG/EEG signals at once is to display only a subset of them at once. This can be done using the "montage" interface. The term refers mainly to EEG, to define the referencing system used to display the values recorded on the electrodes: bipolar montages, custom reference electrodes, average reference. However, the same interface can be used in MEG to select only a subset of sensors in a given figure, using a predefined or custom set of channels. * Use the drop-down menu in the Record tab to several predefined sets (ex. CTF LF = Group of left frontal sensors). Alternatively, right-click on the figure > Montage. You may need to decrease again the gain of the channels (use SHIFT+Mouse wheel)<<BR>><<BR>> {{attachment:selectionsMenu.gif}} * Note the keyboard shortcuts: '''SHIFT+letter'''. It makes it really easy to switch from a group of sensors to another (ex: Shift+A = all sensors, Shift+B = Left-frontal, Shift+C=Right-frontal...) * To edit/create your own selections, use the menu "Edit montages". You can edit the channels selection groups or the electrodes montages available for this installation of Brainstorm. By default, it shows only the ones that are relevant to the type of recordings you are currently looking at (in this case: CTF MEG recordings). To see all the available options, click on the button "All". * Click on "New channel selection" to create a new selection Test1, and select a few channels in the list.<<BR>><<BR>> {{attachment:selections.gif}} * Click on Save. Back to the time series figure. Look in the popup menu again, you can see your new selection in the menu: Test1. You can now use the time series view with these predefined selections of sensors. * To display several montages at the same time, or with different different display options, you need to open twice the same file and then set the parameters differently. You have two option to clone the figure: * Do twice: Right-click on the file in the database explorer > MEG > Display time series * Right-click on a figure > Figure > Clone figure * To have the same scale in both figures, use the buttons "..." in the figures<<BR>><<BR>> {{attachment:figClone.gif}} == Time selection == Go back to the previous view mode ("butterfly view") and in the Record tab, select the menu "All channels" again. Keep this selection for the rest of the tutorial. * Left-click somewhere on the white part of the time series figure, hold the mouse button, and drag your mouse left or right: A transparent blue rectangle appears to represent the time selection. * Now right-click on the figure to see the functions you can apply on this time window. The selected time window and its duration are shown at the bottom of the figure.<<BR>><<BR>> {{attachment:timeSelection.gif}} * '''Set current time''': Just sets the time cursor when the right-click occurred. The shortcut Shift+Click can be useful when trying to move in time on dense displays in columns view. * '''Set selection manually''': Select a time window by giving the bounds in ms. * '''Average time''': Average over the selected time window, saved as a new file in the database. * '''Export to database''': Extract the recordings and save them in a new file in database. * If some sensors are selected, only their values are extracted, all the others are set to zero. * '''Export to file''': Same, but in a user-defined file (not in the database) * '''Export to Matlab''': Same, but export as a Matlab variable in the current workspace. == Bad channels == If you find out that a sensor has unexpected values, that are not coherent with the other surrounding sensors, you can choose to ignore it in the displays and in the source estimation process. In the channel file, each channel has a flag which indicates if it is good or bad. You may also import recordings files that already have some bad channels defined. * Select a few channels with one of the method described above: click on the time series, click on the sensors dots, right-click and move to select a group of sensors. Then right-click in one of the figures and check out the '''Channels''' sub-menu:<<BR>><<BR>> {{attachment:channelMenu.gif}} * '''View selected''': Show the time series of the selected sensors * '''Mark selected as bad''': Remove sensors from the display and all the further computations * '''Mark non-selected as bad''': Keep only the selected channels * '''Reset selection''': Unselect all the selected sensors * '''Mark all channels as good''': Brings back all the channels to display * '''Edit good/bad channels''': Opens an interface that looks like the channel editor, but with one extra column to edit the status (good or bad) of each channel. * '''Mark channels as bad''':<<BR>>Right-click > ''Channels > Mark selected as bad'', or press ''Delete ''key. The sensors should disappear in all figures, and the topography view (2D sensors cap) is updated so that the interpolation on the 2D surface now ignores the bad channels.<<BR>><<BR>> {{attachment:channelSetBad.gif}} * '''Get the channels back''': two options * Right-click on figure > ''Channels > Mark all channels as good<<BR>>'' * Right-click on figure ''> Channels > Edit good/bad channels...'' : this menu open a window very similar to the Channel Editor window introduced in previous tutorials, but without the annoying location and orientation values, and with green and red dots. Click on the dots to mark a channel as good or bad. <<BR>><<BR>>''' {{attachment:channelEditGoodBad.gif}} ''' * Note that if you click on a row in this window, it will select the corresponding channel in the time series and topography figures. * Close this window to save the changes. * '''Batching this from the database explorer''': * You will find a "Channels" menu for any node in the tree that contains recordings. * If you do this on the level of a node, the operation will be applied recursively to all the recordings contained in the node. This way, you can quickly set that the channel 63 is always bad, or that electrode 43 is not working for subject #12, without having to visualize all the recordings one after the other. * The'' View all bad channels'' command displays the list of all the bad channels in all the files in the ''Messages ''tab, in main Brainstorm window. <<BR>><<BR>> {{attachment:channelTreePopup.gif}} * '''Important notes''': * The good/bad channel flags are stored in the recordings files, not in the channel files. So if you marked some channels as ''bad ''in the ''ERF ''data file, there are still considered as ''good ''in the ''Std ''data file. |
|
| Line 20: | Line 268: |
| * Open a ''2D sensor cap'' view, and play with colors. * Brainstorm keeps track of many user-defined colormaps: anatomy, EEG, MEG, sources, stat, time, time-frequency, etc. You can go to the Colormaps menu in the main window to see this list. Usually, you will use only popup menus from specific figures to edit the colormaps. * If you modify a colormap, the changes will be applied to all the figures, saved in your user preferences and available the next time you start Brainstorm. * You are going to manipulate only with the MEG colormap, but the others work exactly the same way. * Right-click on you ''2D sensor cap'' figure > Colormap.<<BR>><<BR>> {{attachment:colormapPopup.gif}} * '''Colormap''': You can change the colors that are used to represent the recorded MEG values. You can create your own colormaps with the Matlab colormap editor, clicking on the last option ''Custom''. * '''Absolute values''': Display the absolute values of the recordings, instead of the original values. * This is the default for ''Anatomy, Sources'' and ''Stat ''colormaps, but it is not very useful for recordings: for EEG and MEG, the sign of the values is very important. <<BR>><<BR>> {{attachment:colormapDefault.gif}} {{attachment:colormapAbsolute.gif}} <<BR>>''__Fig.1__: Relative values; __Fig.2__: Absolute values '' * '''Maximum''': How is the maximum of the colorbar estimated, for the color correspondence. * '''Global''': The bounds of the colormap are set to the maximum value across the whole time window. Eg. if you use the ''rbw'' colormap and the min and max values are [-100ft, +100ft], the colors will be mapped in the following way: -100ft is blue, +100ft is red, 0ft is white. The display is identical for each time sample. If you select this option at t=-49ms, the 2D topography figure will turn almost white because the values are low before the stimulus. * '''Local''': It uses the local min and max values at the current time frame AND for each figure, instead of the global min and max. Eg. at t=-49ms, the extrema values are roughly [-25ft, +25ft]. So the colors will be mapped in order to have: -25ft = blue, and +25ft = red. * '''Custom''': You can set manually the minimum/maximum bounds of the colorbar. It does not have to be symmetrical around zero. If you set the values to [-100, 200] ft, the white colors would correspond to values ~50ft, hence values around 0ft would be displayed in blue.<<BR>><<BR>> {{attachment:colormapNotNormalized.gif}} <<BR>>''__Fig.1__: Local max; __Fig.2__: Global max; __Fig.3__: Custom [-100,300]'' * You can usually keep this option to ''Local'' when looking at recordings, it makes things nicer. But keep in mind that it is not because you see flashy colors that you have strong effects. It's always a matter of colormap configuration. * Remember that when you change this option, it is saved in your user preferences. If you close Brainstorm and start it again, the colormap configuration stays the same. * '''Range''': Use symmetrical or non-symmetrical colormaps. * '''[-max, max]''': Symmetrical colorbar around the absolute value of the maximum. Eg. at t=47ms, the range is [-80ft, +100ft], and the colorbar used is [-100ft, +100ft], white is zero. * '''[min, max]''': Uses the real min and max. Eg. at t=47ms, the colorbar used is [-80ft, +100ft], white is NOT zero. * This option is ignored when the option "Maximum: Custom" is selected.<<BR>><<BR>> {{attachment:colormapMax5.gif}} <<BR>>''__Fig.1__: [-max,max]; __Fig.2__: [min,max] '' * '''Contrast and Brightness''': * Brightness moves the center of the colormap up and down. * Contrast saturate/desaturate the colors.<<BR>><<BR>> {{attachment:colormapContrast.gif}} <<BR>>''__Fig.1__: High brightness; __Fig.2__: High contrast'' * The words brightness/contrast may not be adapted for colormaps such as ''rbw, jet'' or ''hsv. ''It makes more sense for colormaps with only one tint that varies in intensity, such as the ''grey'' colormap. * You can also modify these values by '''clicking directly on the color bar''' in the figures. Hold the mouse button, and move up/down to change the brightness and left/right to change the contrast. Even if the sliders do not work, you can modify the colormaps. * '''Display colorbar''': Just in case you want to hide the color bar... * '''Permanent menu''': Open a window that displays this colormap sub-menu. Might be useful when you do a lot of colormap adjustments. * '''Restore defaults''': Click on it '''now '''so all your experiments will be discarded. * You can also reset the colormap by '''double-clicking on the color bar'''. == Multiple windows == One interesting feature of the Brainstorm interface is to open easily multiple conditions or subjects simultaneously. The buttons in the "Window layout options" menu can help you organize all the opened figures in an efficient way. There are four options for the automatic placement of the figures on the screen and you have the possibility to save your own specific working environment. Open a few figures and try the different options. . {{attachment:toolbarWindows.gif}} ==== Automatic figure positioning ==== * '''Layout options''': Defines how the figures are positioned on the screen * '''Tiled''': All the figures have similar sizes * '''Weighted''': Some figures containing more information are given more space on the screen. This mode is mostly useful when reviewing continuous recordings * '''Full area''': Each figure takes all the space available for figures * '''None''': The new figures are displayed at the default Matlab position, always at the same place, and never re-organized after. Selecting this option can be useful if the auto-arrangement does not work well on your system or if you want to organize your windows by yourself. It is also automatically selected when using "user setups" (see below). * '''One screen / two screens''': If you have multiple monitors, Brainstorm can try to place the database window on one screen and all the other figures on the other screen. If you force Brainstorm to use only one screen, all the figures should stay on the same screen. * '''Full screen''': If selected, the figures are placed using all the screen. If not selected, the figure area is limited not to overlap the Brainstorm window. * '''Show all figures''': If you have many figures hidden by some other fullscreen window (Matlab, Firefox to read this tutorial, etc.), you don't have click on all of them in the taskbar to get them back. Just make the Brainstorm window visible and click on this button, it would bring all the figures back (not working on some Linux window managers). * '''User setups''': You can save a combination of figures currently opened on your desktop and re-use it later on a different dataset. It may not look that interesting when working with averaged files, but it becomes very useful when reviewing numerous and large continuous files. * '''Close all windows''': Close everything and free most of the allocated memory. * Open multiple figures and observe the behavior of the different layout options.<<BR>><<BR>> {{attachment:figuresNotTiled.gif}} -- {{attachment:figuresTiled.gif}} ==== User setups ==== * Open many views on one unique data file, ''Left / ERF'', for instance: * MEG time series "butterfly" * MEG time series "columns" with only the left-temporal sensors * 2D sensor cap * 3D sensor cap * Re-arrange the windows the way you want on the screen * In the ''Window layout'' menu, select User setups > New setup > "TestCTF". * Note that there is a new entry in ''User setups'' and that the ''None'' layout is now selected. Using custom window configurations disables the automatic arrangement of the windows on the screen. * Close all the figures. * Double-click on ''Right / ERF'' to open it. * Select the menu User setups > TestCTF. It should re-open all the figures that were previously defined for the Left/ERF file, organized in the way you saved them.<<BR>><<BR>> {{attachment:userSetupLeft.gif|figuresNotTiled.gif}} -- {{attachment:userSetupRight.gif|figuresNotTiled.gif}} ==== Uniform amplitude scales ==== * The button ''Uniform amplitude scale'' {{attachment:buttonEqual.gif|buttonTsCol.gif}} in the Record tab can be useful when working with multiple time series figures. If this option is selected, all the time series figures with similar units have the same y-axis scale, so that you can compare visually the amplitudes between two datasets. * Double-click on ''Right / ERF'' and ''Left / Std'', and click on this button several times. * The scale of the amplitude axis changes in the ''Left/Std ''window, and fit alternatively its own maximum or the maximum in the other figure.<<BR>><<BR>>Uniform amplitude on:<<BR>> {{attachment:figuresUniform.gif}} <<BR>><<BR>>Uniform amplitude off:<<BR>> {{attachment:figuresNotUniform.gif}} == Database navigator == The Navigator menu can help you to go quickly from a dataset to another. It can be almost indispensable when your are reviewing 200 trials of the same MEG response. {{attachment:navigator.gif}} * You can access it from the popup menus of all the figures showing functional data. * You can also use the keyboard shortcuts:'' F1, F2, F3'', together with ''Shift ''key to go backwards. * Now, close all the figures (use the ''Close all figures ''button) * For ''Right / ERF'', display three views : * Time series (double click) * 2D sensor cap (Ctrl+T)''' ''' * 2D Layout (right click on ERF file > Display > 2D Layout) * Press ''F3 ''once: Updates all the figures to display ''Right/Std'' and selects it in the database explorer * Press ''F3 ''again: Nothing happens, you are already at the last dataset for this subject / condition. * Press ''Shift + F3'': And you'll be back to ''ERF ''file. * Press ''F2'': Similar to F3, but jumps from a condition to another, within the same subject. * If you had many subjects you could also use ''F1 / Shift+F1''. * __MacBook users__: The keys "Fx" are obtained by holding the "Fn" key simultaneously. == Snapshots == Using Brainstorm, you will quickly feel like saving the beautiful images you produce. For that you can: * Press the ''PrintScreen'' key on your keyboard and paste the copied screen in your favorite image or text editor (PC or Linux) * More subtle: ''Alt+PrintScreen'' would only copy the figure that is currently selected (PC or Linux). * On MacOS: Many more options available, Google for the best ones ([[http://graphicssoft.about.com/od/screencapturemac/ht/macscreenshot.htm|see example]]). * The ''Snapshots ''menu present in the popup of all the figures can be useful too. Here are two examples of Snapshots menus, respectively on time series and 3D figures:<<BR>><<BR>> {{attachment:snapshotsTS.gif}} --- {{attachment:snapshotsTopo.gif}} * '''Save as image''': save the figure (without the title bar and borders) in a file. Many formats available. * '''Open as image''': capture the figure and open it in as an image. This can be useful if you want to compare visually the selected figure with another one that you cannot display at the same time (because they have different time or frequency definitions) * '''Export to database''': saves the recordings in the figure as a new entry in database * If there are selected channels, only their values will be saved, the others being set to zero. * '''Save time series''': extract the time series displayed in this figure (or only the selected sensors), and save them in a file. Available formats: * ASCII (.txt) * Matlab (.mat): saves much more information (titles, time values, etc.) * Cartool (.eph) * EGI (.raw) * '''Export to Matlab''': Same thing, but exports the structure in a variable in Matlab workspace instead of creating a new file. * '''Movie (time): Selected figure''': Create .avi movies to show time evolution of the selected figure. * The dimensions of the movie image depend on the actual size of the figure on the screen. * Don't do anything else while creating the movies: the figure which is captured must be visible all the time. * '''Movie (time): All figures''': Instead of capturing one figure only, it captures them all and creates a movie showing what you see on the screen. Arrange your figures the way you want and create a movie of all your workspace at once.<<BR>><<BR>> {{attachment:movie.gif}} * '''Movie (horizontal/vertical)''': Rotate spatially the 3D scene. * '''Contact sheet''': Produce a big image with all the time frames in it. * Same recommendations than for movies: if you don't want the final image to be too big, reduce the size of your figure, zoom a bit, and maybe hide the colorbar. Don't hide the figure during the capture. * At the end, the image is displayed in a viewer with which you can zoom (menu or wheel), move (left-click+move), and save the image (File > Save as). * The contact sheet menu also appears for the time series figure, which is not that interesting because nothing changes except for the position of the time cursor. However, it can make sense to use in combination with the same contact sheet for a 2D/3D figure. Displaying the two images side by side you would have a clear view of the exact timing of each topography. <<BR>><<BR>> {{attachment:contactSheet.gif||height="299px",width="487px"}} * '''Figure menu''': Various options to edit the selected figure. == Keyboard shortcuts == Here is a memo of all the keyboard shortcuts for time series and topography figures. If you don't remember them, you can find most of them in the figure popup menus. * '''Arrows: '''left, right, PageUp, PageDown: Move in time * '''Delete''': Mark selected sensors as ''bad'' * '''Shift + ''''''Delete''': Mark non-selected sensors as ''bad'' (=keeps ony the selected sensors) * '''Enter''': View time series for selected sensors * '''Shift + Enter''': Set all the bad sensors as good (=brings back all the channels in the display) * '''Escape''': Unselect all the selected sensors * '''Ctrl + A''': Show axis on 3D figures (X,Y,Z) * '''Ctrl + A''': Add event in raw file viewer (see raw file viewer tutorial) * '''Ctrl + B''': Set trial as bad * '''Ctrl + D''': Dock/undock figure in Matlab's figures list * '''Ctrl + E''': Show sensors and labels (E stands initially for ''Electrode'') * '''Ctrl + I''': Save figure as image * '''Ctrl + R''': Open ''Time series'' view (R stands for ''Recordings'') * '''Ctrl + S''': Open ''Sources'' view (see next tutorial) * '''Ctrl + T''': Open ''2D sensor cap'' view (T stands for ''Topography'') * '''Shift + letter''': Change sensors display when in "column" display mode for the time series. * '''F1, F2, F3''': with or without '''Shift''', calls the database navigator (F1=subject, F2=condition, F3=file) * __Notes for Mac users__: * PageUp = Fn + UP * PageDown = Fn + DOWN * F1 = Fn + F1 * Mouse wheel = Two finger up/down on the MacBook pad == Next == Now you are able to use all the tools that are available for importing and displaying your MEG or EEG recordings, and should start to feel comfortable with the database explorer.The next two steps are the resolution of the forward problem and the inverse problem. [[Tutorials/TutHeadModel|Next]]. <<EmbedContent(http://neuroimage.usc.edu/brainstorm3_register/get_feedback.php?Tutorials/TutExploreRecodings)>> |
[WARNING: This page is part of the old tutorials, please refer to the new documentation]
Tutorial 5: Exploring the recordings
Authors: Francois Tadel, John C Mosher, Sylvain Baillet
This tutorial is based on the TutorialCTF protocol, and will show you how to explore efficiently the sensory response to an electric stimulation of the fingers.
Contents
Display recordings
Several display modes are available for recordings. They are all accessible with a right-click.The first four menus (MEG, MEG REF, Stim, Video), represent all the different types of channels that were found in the channel file. You can check that with Channel Edit (right-click on channel file > Edit). We are only interested in the "MEG" in this tutorial. Select Left / ERF > MEG > Display time series.
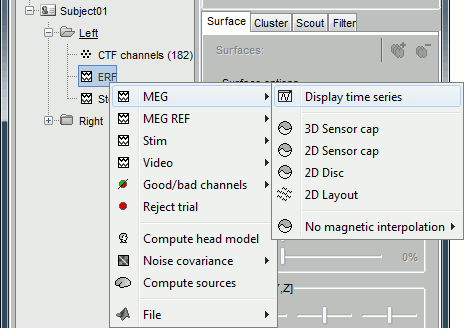
Now repeat this operation several times to display all the available modes: Display time series, 3D sensor cap, 2D sensor cap, 2D disc and 2D Layout. You should now see all the figures displayed below. Keep them opened for the next few paragraphs, you will learn how to manipulate multiple views of the same data.
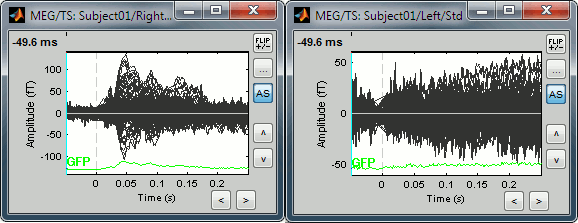
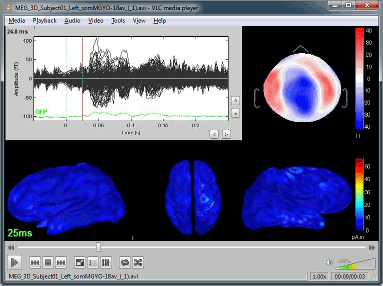
Display time series
Amplitude fluctuations over time for all the sensors.
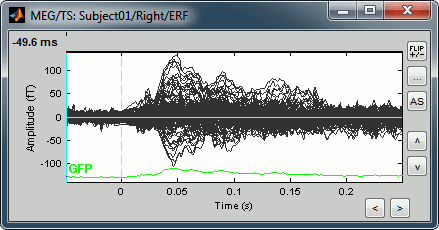
Buttons available on the figure:
[<] and [>]: Horizontal zoom around the current time point
[^] and [v]: Vertical zoom
[Flip +/-]: Exchange the direction of the Y axis, useful in clinical EEG
[...]: Set the amplitude range manually, ie. the limits of the Y axis
[AS]: Amplitude auto-scale. When selected, the amplitude range (Y axis limits) is automatically updated when the figure is updated. This is useful mostly when reviewing continuous files or individual trials, it will be illustrated in other tutorials.
List of mouse and keyboard operations for this type of figure (most of the operations are detailed later):
Mouse wheel: Horizontal zoom
CTRL+mouse wheel: Vertical zoom
Right click + move: Move in zoomed figure
Left click + move: Time selection
Left-click: Set current time
Left-click on a line: Select the corresponding sensor
Shift + Left-click: Force setting the current time, even when clicking on a sensor
Right-click: Popup menu
Double-click: Restore initial view (ie. unzoom)
Keyboard arrows: Change current time
PageUp / PageDown: Change current time (10 samples at a time)
Notes for Mac users:
All the actions involving the mouse wheel can be performed on MacBook laptops by moving up and down two fingers on the pad.
"PageUp" or "PageDown" keys are obtained by pressing the "Fn" key together with the arrows UP or DOWN
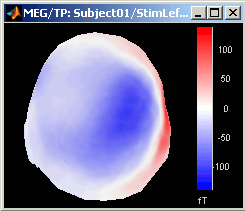
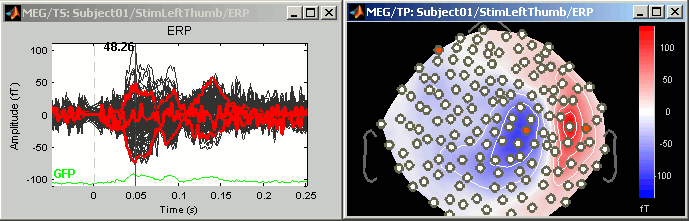
3D Sensor cap / 2D Sensor cap / 2D Disc
The recordings at a given time instant and interpolated over a 2D or 3D surface.
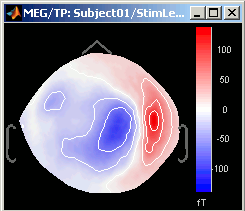
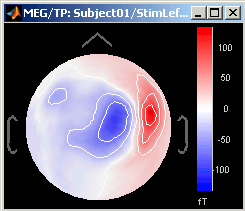
Keyboard and mouse shortcuts:
Mouse wheel: Zoom / unzoom
Left click + move: Rotate (3D only)
Middle click + move: Or left+right click + move: Move in zoomed figure
Right click + move: Select sensors (2D only, when sensors are visible, see below)
Right click: Popup menu
Left click on the colorbar + move: Change contrast (up/down) and brightness (left/right)
Control + E: Display the channels markers and/or names
Keyboard arrows: Change current time
PageUp / PageDown: Change current time (10 samples at a time) / Mac: Fn+UP/Fn+Down
2D layout
The time course of each channel is drawn at the actual position of the electrode, projected in 2D the same way as 2D Sensor cap. The light gray lines represent the zero amplitude (horizontal) and the current time (vertical lines).
Only a part of the full time window is displayed for each channel, before and after the current time. The length of this time window can be modified either with the mouse shortcut Control+wheel, or with the 2D Layout options, in the figure popup menu. Other options are also available in the popup menu.
Mouse wheel: Zoom / unzoom
Middle click + move: (or left+right click + move) Move into the zoomed figure
Shift + mouse wheel: Increase/decrease the amplitude gain of the channels
Control + mouse wheel: Increase/decrease the length of the time window around the current time
Click on a line: Select a sensor
Right click + move: Select a group of sensors
Shift + click on a line: Select one sensor and unselect all the others
Display the time-frequency decomposition for the selected sensor, when availableRight-click: Display popup menu.
Sub-menu "2DLayout options" offer some configuration options for this type of figure.Control + E: Display/hide the channels names
Keyboard arrows: Change current time
PageUp / PageDown: Change current time (10 samples at a time) / Mac: Fn+UP/Fn+Down
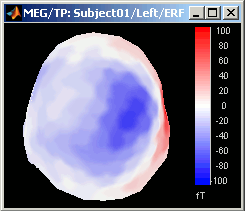
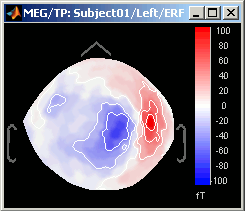
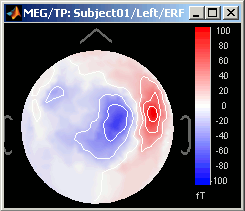
Magnetic interpolation
Some of the views (3D Sensor cap / 2D Sensor cap / 2D Disc), are by default re-interpolating the field that is recorded by the sensors to get much smoother displays. A simple inverse problem + forward problem are solved to reconstruct the magnetic fields on a high-resolution surface.
The menu "No magnetic interpolation" offer the same views, but without using this reconstruction of the magnetic field, and performing instead a spatial interpolation of the values between the sensors.
Graphic bugs
If you observe any graphic problem with these displays, there might be an issue with the OpenGL drivers. You may try the following options:
- Update the drivers for your graphics card.
- Upgrade your version of Matlab.
Run the compiled version of Brainstorm (see Installation).
Turn off the OpenGL hardware acceleration: Menu File > Edit preferences > Software or Disabled.
- Send a bug report to the Mathworks.
Time exploration
The time window and the current time instant are centralized and managed from the top of the Brainstorm window. You can use the text box or the buttons to change the current time value. All the figures always stay synchronized, you cannot display the 2D disc topography at t=10ms and the 3D sensor cap topography at t=50ms at the same time. It might look too restrictive at the beginning, but the contrary would have made the interface much too complex; we wanted to keep it as simple and intuitive as possible.
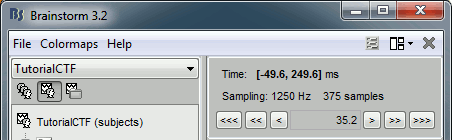
The time window panel is not the best way to explore your recordings. There are many other shortcuts that are much faster to navigate through the time.
Click on time series figure:
Left-click in any white area of the figure: it will move the time cursor at this point and update all the other figures to match this new value. Don't click on a black line, it would select the corresponding channel instead of moving the time cursor.Press the keyboard arrows:
Click on any data figure and try to press several time the left and right arrows, and also the PageUp and PageDown keys. This feature is really useful when you want to quickly see how the recordings are changing in time. Try holding the arrow keys for a while. It might be a bit slow because you have many figures to refresh at each time change. It is much faster if you have only one 2D figure to refresh.
To remember: An efficient and quick way to review your recordings just after importation is to:
- Open a Time series window (double click on recordings file)
- Click on the time of interest (eg. first response peak)
Open a topography view of your choice (2D disc, 2D sensor cap, or 3D sensor cap)
You can do this by right-clicking on the time series figure > MEG Topography (or Ctrl+T keyboard shortcut).Use the left/right arrows, and PageUp/PageDown keys (or Fn+Up / Fn+Down on a Mac)
Sensors selection
You can select some channels, and then display them separately or mark them as good or bad.
Now close all the figures except the Time series and the 2D Sensor cap topography (if you don't remember which one it was, close everything and open again these two figures).
On 2D Sensor cap figure: right-click anywhere on the window > Channels > Display sensors. You'll see white dots representing the center of each MEG coil. Note that there is keyboard shortcut indicated for this menu: Ctrl + E.
Press Ctrl + E several times and see what happens. Come back to the display with only the sensors markers (no labels).
- Click on some white dots.
- They turn red and the corresponding lines in the time series window also turn red
- Left-click on the lines in the time series window: it also selects the sensors
- Right-click on a red line in time series figure: shows the name and index of the selected channel
- Click on a selected (red) point or line: the sensor is deselected.
To deselect all the sensors, press Escape or right-click > Channels > Reset selection
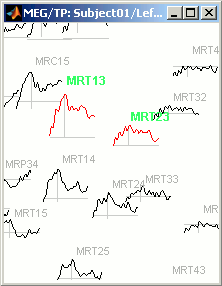
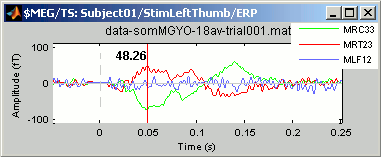
Now select randomly three sensors.
Right-click > Channels > View selected (Shortcut = Enter key). A new window is created, with only these three sensors, whose names are indicated in a legend box (you can move this legend if needed).
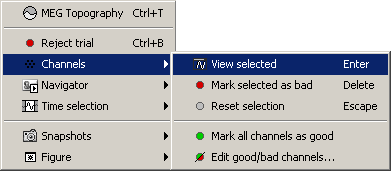
Close that last figure. In the 2D topography figure, right-click and move the mouse to select a group of sensors, just to remember that this feature exists.
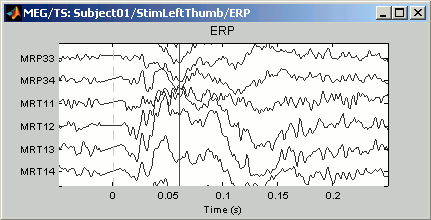
Time series in columns
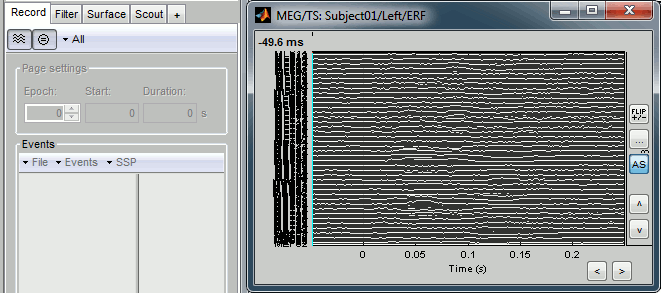
There are two display modes for the MEG signals: "butterfly", what we've seen until now, or "column". Double click on Left/ERP to open a time series figure. To switch from the current mode to the "column" mode, click on  in the Record tab.
in the Record tab.
The figure you get is unreadable because it contains the 151 sensors in the same figure. There are several ways to get a readable display: you need to select a subset of the channels, zoom and/or to increase the gain of the channels.
- Start by maximizing the size of the figure. If you have a large screen, you might be already able to see something.
Channel gain: You can increase/decrease the amplitude of the traces in different ways:
- Click on the up/down buttons on the right side of the figure
- Click on the "..." button on the right of the window to set manually the gain value for each vertical unit in the graph (distance between two horizontal lines).
- Press the "+" or "-" keys on your keyboard
- Hold the SHIFT key down, and use the mouse wheel to zoom/unzoom
Vertical zoom: Hold the CTRL key down, and use the mouse wheel to zoom in or out.
Horizontal zoom:
Click on the "<" and ">" buttons at the bottom of the window
- Use the mouse wheel to zoom in or out.
Move in the zoomed figure: Hold down left and right buttons of the mouse, and move the mouse.
You can reach some display results that are already a little more interesting:
Reset initial view: Double-click
Montages
A better way to review a large number of MEG/EEG signals at once is to display only a subset of them at once. This can be done using the "montage" interface. The term refers mainly to EEG, to define the referencing system used to display the values recorded on the electrodes: bipolar montages, custom reference electrodes, average reference. However, the same interface can be used in MEG to select only a subset of sensors in a given figure, using a predefined or custom set of channels.
Use the drop-down menu in the Record tab to several predefined sets (ex. CTF LF = Group of left frontal sensors). Alternatively, right-click on the figure > Montage. You may need to decrease again the gain of the channels (use SHIFT+Mouse wheel)
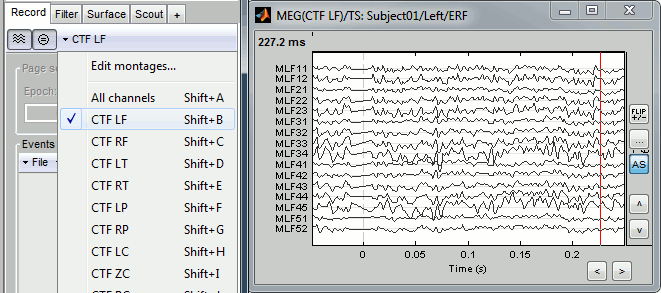
Note the keyboard shortcuts: SHIFT+letter. It makes it really easy to switch from a group of sensors to another (ex: Shift+A = all sensors, Shift+B = Left-frontal, Shift+C=Right-frontal...)
- To edit/create your own selections, use the menu "Edit montages". You can edit the channels selection groups or the electrodes montages available for this installation of Brainstorm. By default, it shows only the ones that are relevant to the type of recordings you are currently looking at (in this case: CTF MEG recordings). To see all the available options, click on the button "All".
Click on "New channel selection" to create a new selection Test1, and select a few channels in the list.
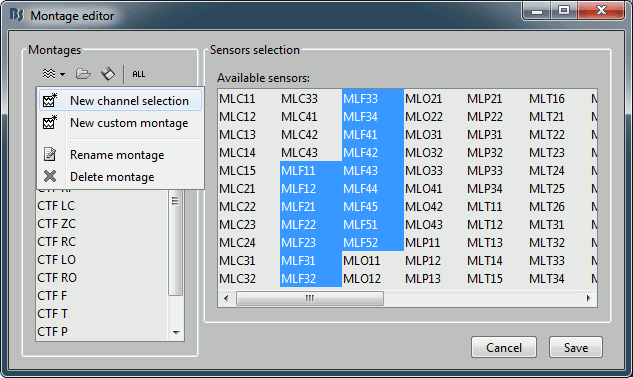
- Click on Save. Back to the time series figure. Look in the popup menu again, you can see your new selection in the menu: Test1. You can now use the time series view with these predefined selections of sensors.
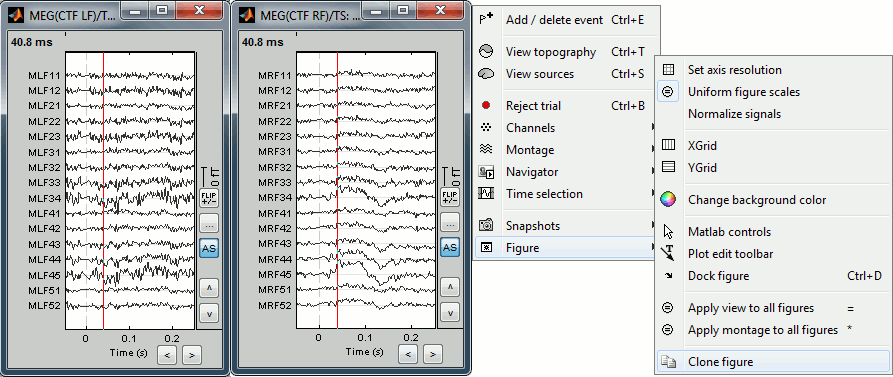
- To display several montages at the same time, or with different different display options, you need to open twice the same file and then set the parameters differently. You have two option to clone the figure:
Do twice: Right-click on the file in the database explorer > MEG > Display time series
Right-click on a figure > Figure > Clone figure
To have the same scale in both figures, use the buttons "..." in the figures
Time selection
Go back to the previous view mode ("butterfly view") and in the Record tab, select the menu "All channels" again. Keep this selection for the rest of the tutorial.
- Left-click somewhere on the white part of the time series figure, hold the mouse button, and drag your mouse left or right: A transparent blue rectangle appears to represent the time selection.
Now right-click on the figure to see the functions you can apply on this time window. The selected time window and its duration are shown at the bottom of the figure.
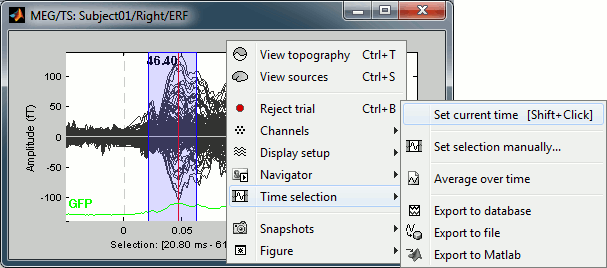
Set current time: Just sets the time cursor when the right-click occurred. The shortcut Shift+Click can be useful when trying to move in time on dense displays in columns view.
Set selection manually: Select a time window by giving the bounds in ms.
Average time: Average over the selected time window, saved as a new file in the database.
Export to database: Extract the recordings and save them in a new file in database.
- If some sensors are selected, only their values are extracted, all the others are set to zero.
Export to file: Same, but in a user-defined file (not in the database)
Export to Matlab: Same, but export as a Matlab variable in the current workspace.
Bad channels
If you find out that a sensor has unexpected values, that are not coherent with the other surrounding sensors, you can choose to ignore it in the displays and in the source estimation process. In the channel file, each channel has a flag which indicates if it is good or bad. You may also import recordings files that already have some bad channels defined.
Select a few channels with one of the method described above: click on the time series, click on the sensors dots, right-click and move to select a group of sensors. Then right-click in one of the figures and check out the Channels sub-menu:
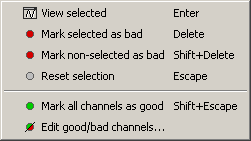
View selected: Show the time series of the selected sensors
Mark selected as bad: Remove sensors from the display and all the further computations
Mark non-selected as bad: Keep only the selected channels
Reset selection: Unselect all the selected sensors
Mark all channels as good: Brings back all the channels to display
Edit good/bad channels: Opens an interface that looks like the channel editor, but with one extra column to edit the status (good or bad) of each channel.
Mark channels as bad:
Right-click > Channels > Mark selected as bad, or press Delete key. The sensors should disappear in all figures, and the topography view (2D sensors cap) is updated so that the interpolation on the 2D surface now ignores the bad channels.
Get the channels back: two options
Right-click on figure > Channels > Mark all channels as good
Right-click on figure > Channels > Edit good/bad channels... : this menu open a window very similar to the Channel Editor window introduced in previous tutorials, but without the annoying location and orientation values, and with green and red dots. Click on the dots to mark a channel as good or bad.
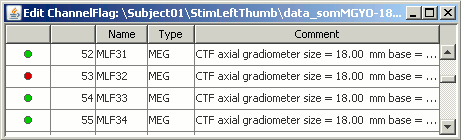
- Note that if you click on a row in this window, it will select the corresponding channel in the time series and topography figures.
- Close this window to save the changes.
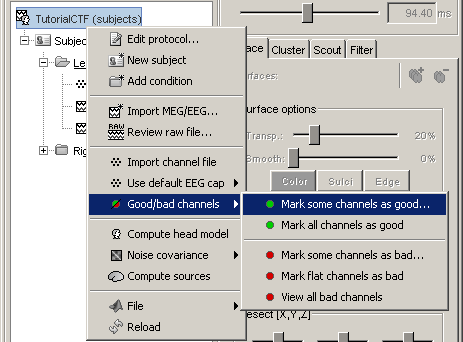
Batching this from the database explorer:
- You will find a "Channels" menu for any node in the tree that contains recordings.
- If you do this on the level of a node, the operation will be applied recursively to all the recordings contained in the node. This way, you can quickly set that the channel 63 is always bad, or that electrode 43 is not working for subject #12, without having to visualize all the recordings one after the other.
The View all bad channels command displays the list of all the bad channels in all the files in the Messages tab, in main Brainstorm window.
Important notes:
The good/bad channel flags are stored in the recordings files, not in the channel files. So if you marked some channels as bad in the ERF data file, there are still considered as good in the Std data file.
Colormap configuration
Open a 2D sensor cap view, and play with colors.
- Brainstorm keeps track of many user-defined colormaps: anatomy, EEG, MEG, sources, stat, time, time-frequency, etc. You can go to the Colormaps menu in the main window to see this list. Usually, you will use only popup menus from specific figures to edit the colormaps.
- If you modify a colormap, the changes will be applied to all the figures, saved in your user preferences and available the next time you start Brainstorm.
- You are going to manipulate only with the MEG colormap, but the others work exactly the same way.
Right-click on you 2D sensor cap figure > Colormap.
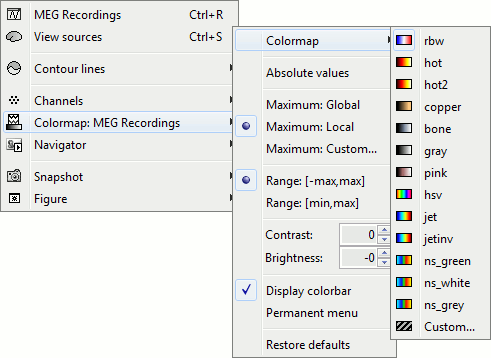
Colormap: You can change the colors that are used to represent the recorded MEG values. You can create your own colormaps with the Matlab colormap editor, clicking on the last option Custom.
Absolute values: Display the absolute values of the recordings, instead of the original values.
This is the default for Anatomy, Sources and Stat colormaps, but it is not very useful for recordings: for EEG and MEG, the sign of the values is very important.
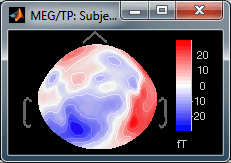
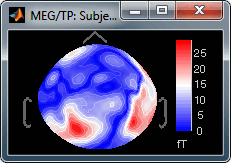
Fig.1: Relative values; Fig.2: Absolute values
Maximum: How is the maximum of the colorbar estimated, for the color correspondence.
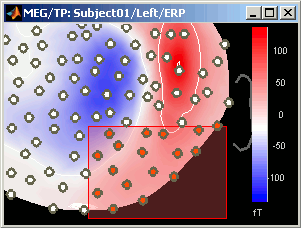
Global: The bounds of the colormap are set to the maximum value across the whole time window. Eg. if you use the rbw colormap and the min and max values are [-100ft, +100ft], the colors will be mapped in the following way: -100ft is blue, +100ft is red, 0ft is white. The display is identical for each time sample. If you select this option at t=-49ms, the 2D topography figure will turn almost white because the values are low before the stimulus.
Local: It uses the local min and max values at the current time frame AND for each figure, instead of the global min and max. Eg. at t=-49ms, the extrema values are roughly [-25ft, +25ft]. So the colors will be mapped in order to have: -25ft = blue, and +25ft = red.
Custom: You can set manually the minimum/maximum bounds of the colorbar. It does not have to be symmetrical around zero. If you set the values to [-100, 200] ft, the white colors would correspond to values ~50ft, hence values around 0ft would be displayed in blue.
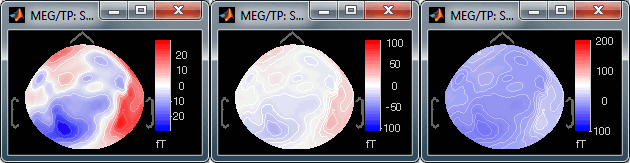
Fig.1: Local max; Fig.2: Global max; Fig.3: Custom [-100,300]You can usually keep this option to Local when looking at recordings, it makes things nicer. But keep in mind that it is not because you see flashy colors that you have strong effects. It's always a matter of colormap configuration.
- Remember that when you change this option, it is saved in your user preferences. If you close Brainstorm and start it again, the colormap configuration stays the same.
Range: Use symmetrical or non-symmetrical colormaps.
[-max, max]: Symmetrical colorbar around the absolute value of the maximum. Eg. at t=47ms, the range is [-80ft, +100ft], and the colorbar used is [-100ft, +100ft], white is zero.
[min, max]: Uses the real min and max. Eg. at t=47ms, the colorbar used is [-80ft, +100ft], white is NOT zero.
This option is ignored when the option "Maximum: Custom" is selected.
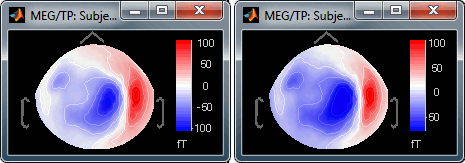
Fig.1: [-max,max]; Fig.2: [min,max]
Contrast and Brightness:
- Brightness moves the center of the colormap up and down.
Contrast saturate/desaturate the colors.
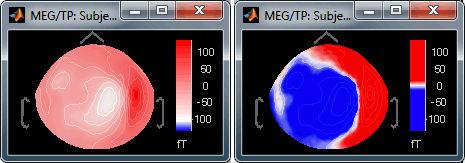
Fig.1: High brightness; Fig.2: High contrastThe words brightness/contrast may not be adapted for colormaps such as rbw, jet or hsv. It makes more sense for colormaps with only one tint that varies in intensity, such as the grey colormap.
You can also modify these values by clicking directly on the color bar in the figures. Hold the mouse button, and move up/down to change the brightness and left/right to change the contrast. Even if the sliders do not work, you can modify the colormaps.
Display colorbar: Just in case you want to hide the color bar...
Permanent menu: Open a window that displays this colormap sub-menu. Might be useful when you do a lot of colormap adjustments.
Restore defaults: Click on it now so all your experiments will be discarded.
You can also reset the colormap by double-clicking on the color bar.
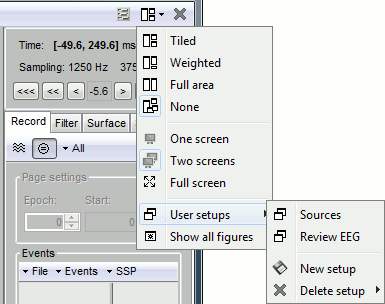
Multiple windows
One interesting feature of the Brainstorm interface is to open easily multiple conditions or subjects simultaneously. The buttons in the "Window layout options" menu can help you organize all the opened figures in an efficient way. There are four options for the automatic placement of the figures on the screen and you have the possibility to save your own specific working environment. Open a few figures and try the different options.
Automatic figure positioning
Layout options: Defines how the figures are positioned on the screen
Tiled: All the figures have similar sizes
Weighted: Some figures containing more information are given more space on the screen. This mode is mostly useful when reviewing continuous recordings
Full area: Each figure takes all the space available for figures
None: The new figures are displayed at the default Matlab position, always at the same place, and never re-organized after. Selecting this option can be useful if the auto-arrangement does not work well on your system or if you want to organize your windows by yourself. It is also automatically selected when using "user setups" (see below).
One screen / two screens: If you have multiple monitors, Brainstorm can try to place the database window on one screen and all the other figures on the other screen. If you force Brainstorm to use only one screen, all the figures should stay on the same screen.
Full screen: If selected, the figures are placed using all the screen. If not selected, the figure area is limited not to overlap the Brainstorm window.
Show all figures: If you have many figures hidden by some other fullscreen window (Matlab, Firefox to read this tutorial, etc.), you don't have click on all of them in the taskbar to get them back. Just make the Brainstorm window visible and click on this button, it would bring all the figures back (not working on some Linux window managers).
User setups: You can save a combination of figures currently opened on your desktop and re-use it later on a different dataset. It may not look that interesting when working with averaged files, but it becomes very useful when reviewing numerous and large continuous files.
Close all windows: Close everything and free most of the allocated memory.
Open multiple figures and observe the behavior of the different layout options.
 --
-- 
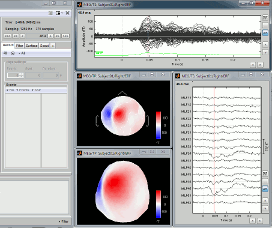
User setups
Open many views on one unique data file, Left / ERF, for instance:
- MEG time series "butterfly"
- MEG time series "columns" with only the left-temporal sensors
- 2D sensor cap
- 3D sensor cap
- Re-arrange the windows the way you want on the screen
In the Window layout menu, select User setups > New setup > "TestCTF".
Note that there is a new entry in User setups and that the None layout is now selected. Using custom window configurations disables the automatic arrangement of the windows on the screen.
- Close all the figures.
Double-click on Right / ERF to open it.
Select the menu User setups > TestCTF. It should re-open all the figures that were previously defined for the Left/ERF file, organized in the way you saved them.
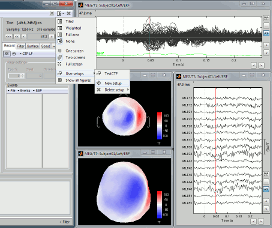 --
-- 
Uniform amplitude scales
The button Uniform amplitude scale
 in the Record tab can be useful when working with multiple time series figures. If this option is selected, all the time series figures with similar units have the same y-axis scale, so that you can compare visually the amplitudes between two datasets.
in the Record tab can be useful when working with multiple time series figures. If this option is selected, all the time series figures with similar units have the same y-axis scale, so that you can compare visually the amplitudes between two datasets. Double-click on Right / ERF and Left / Std, and click on this button several times.
The scale of the amplitude axis changes in the Left/Std window, and fit alternatively its own maximum or the maximum in the other figure.
Uniform amplitude on:
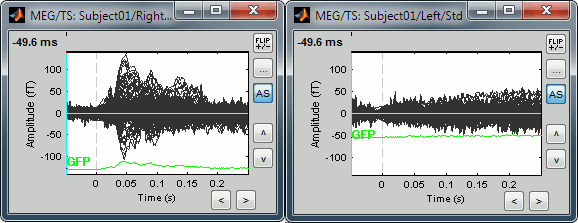
Uniform amplitude off:
Database navigator
The Navigator menu can help you to go quickly from a dataset to another. It can be almost indispensable when your are reviewing 200 trials of the same MEG response.
- You can access it from the popup menus of all the figures showing functional data.
You can also use the keyboard shortcuts: F1, F2, F3, together with Shift key to go backwards.
Now, close all the figures (use the Close all figures button)
For Right / ERF, display three views :
- Time series (double click)
2D sensor cap (Ctrl+T)
2D Layout (right click on ERF file > Display > 2D Layout)
Press F3 once: Updates all the figures to display Right/Std and selects it in the database explorer
Press F3 again: Nothing happens, you are already at the last dataset for this subject / condition.
Press Shift + F3: And you'll be back to ERF file.
Press F2: Similar to F3, but jumps from a condition to another, within the same subject.
If you had many subjects you could also use F1 / Shift+F1.
MacBook users: The keys "Fx" are obtained by holding the "Fn" key simultaneously.
Snapshots
Using Brainstorm, you will quickly feel like saving the beautiful images you produce. For that you can:
Press the PrintScreen key on your keyboard and paste the copied screen in your favorite image or text editor (PC or Linux)
More subtle: Alt+PrintScreen would only copy the figure that is currently selected (PC or Linux).
On MacOS: Many more options available, Google for the best ones (see example).
The Snapshots menu present in the popup of all the figures can be useful too. Here are two examples of Snapshots menus, respectively on time series and 3D figures:
 ---
--- 
Save as image: save the figure (without the title bar and borders) in a file. Many formats available.
Open as image: capture the figure and open it in as an image. This can be useful if you want to compare visually the selected figure with another one that you cannot display at the same time (because they have different time or frequency definitions)
Export to database: saves the recordings in the figure as a new entry in database
- If there are selected channels, only their values will be saved, the others being set to zero.
Save time series: extract the time series displayed in this figure (or only the selected sensors), and save them in a file. Available formats:
- ASCII (.txt)
- Matlab (.mat): saves much more information (titles, time values, etc.)
- Cartool (.eph)
- EGI (.raw)
Export to Matlab: Same thing, but exports the structure in a variable in Matlab workspace instead of creating a new file.
Movie (time): Selected figure: Create .avi movies to show time evolution of the selected figure.
- The dimensions of the movie image depend on the actual size of the figure on the screen.
- Don't do anything else while creating the movies: the figure which is captured must be visible all the time.
Movie (time): All figures: Instead of capturing one figure only, it captures them all and creates a movie showing what you see on the screen. Arrange your figures the way you want and create a movie of all your workspace at once.
Movie (horizontal/vertical): Rotate spatially the 3D scene.
Contact sheet: Produce a big image with all the time frames in it.
- Same recommendations than for movies: if you don't want the final image to be too big, reduce the size of your figure, zoom a bit, and maybe hide the colorbar. Don't hide the figure during the capture.
At the end, the image is displayed in a viewer with which you can zoom (menu or wheel), move (left-click+move), and save the image (File > Save as).
The contact sheet menu also appears for the time series figure, which is not that interesting because nothing changes except for the position of the time cursor. However, it can make sense to use in combination with the same contact sheet for a 2D/3D figure. Displaying the two images side by side you would have a clear view of the exact timing of each topography.
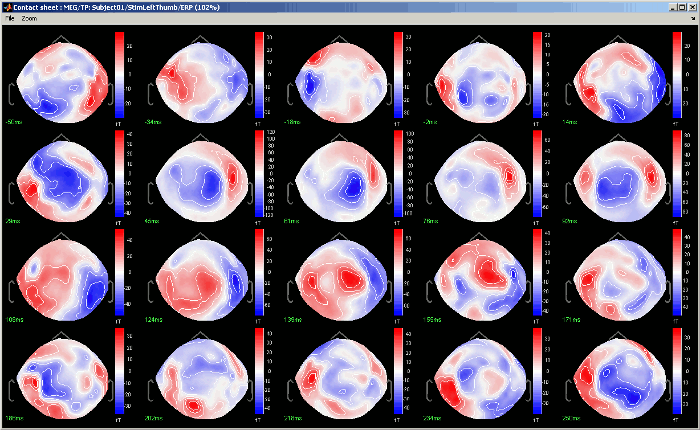
Figure menu: Various options to edit the selected figure.
Keyboard shortcuts
Here is a memo of all the keyboard shortcuts for time series and topography figures. If you don't remember them, you can find most of them in the figure popup menus.
Arrows: left, right, PageUp, PageDown: Move in time
Delete: Mark selected sensors as bad
Shift + Delete: Mark non-selected sensors as bad (=keeps ony the selected sensors)
Enter: View time series for selected sensors
Shift + Enter: Set all the bad sensors as good (=brings back all the channels in the display)
Escape: Unselect all the selected sensors
Ctrl + A: Show axis on 3D figures (X,Y,Z)
Ctrl + A: Add event in raw file viewer (see raw file viewer tutorial)
Ctrl + B: Set trial as bad
Ctrl + D: Dock/undock figure in Matlab's figures list
Ctrl + E: Show sensors and labels (E stands initially for Electrode)
Ctrl + I: Save figure as image
Ctrl + R: Open Time series view (R stands for Recordings)
Ctrl + S: Open Sources view (see next tutorial)
Ctrl + T: Open 2D sensor cap view (T stands for Topography)
Shift + letter: Change sensors display when in "column" display mode for the time series.
F1, F2, F3: with or without Shift, calls the database navigator (F1=subject, F2=condition, F3=file)
Notes for Mac users:
PageUp = Fn + UP
PageDown = Fn + DOWN
- F1 = Fn + F1
Mouse wheel = Two finger up/down on the MacBook pad
Next
Now you are able to use all the tools that are available for importing and displaying your MEG or EEG recordings, and should start to feel comfortable with the database explorer.The next two steps are the resolution of the forward problem and the inverse problem. ?Next.