|
Size: 13135
Comment:
|
Size: 10610
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| This tutorial uses Sabine Meunier's somatotopy experiment, called ''TutorialCTF ''in your Brainstorm database. | The main window includes a graphical batching interface that directly benefits from the database explorer: files are organized as a tree of subjects and conditions, and simple drag-and-drop operations readily select files for subsequent batch processing. Most of the Brainstorm features are (or will be) available through this interface, including pre-processing of the recordings, averaging, time-frequency decompositions, and computing statistics. A full analysis pipeline can be created in a few minutes, saved in the user’s preferences and reloaded in one click, executed directly or exported as a Matlab script. |
| Line 4: | Line 4: |
| The ''Processes ''tab in Brainstorm window is a very generic tool that can be used to apply a function or extract data from any set of data files (recordings or sources). Its main applications are: pre-processing, averaging, and data extraction. | The available processes are organized in a plug-in structure. Any Matlab script that is added to the plug-in folder (brainstorm3/toolbox/process/functions/) and has the right format will be automatically detected and made available in the GUI. This mechanism makes the contribution from other developers to Brainstorm very easy. |
| Line 6: | Line 6: |
| <<TableOfContents(2)>> | == Selecting files to process == First thing to do is to define the files you are going to process. This is done easily by picking files or folders in the database explorer and droping them in the empty list of the Process1 tab. |
| Line 8: | Line 9: |
| == Files to process == First, you have to select the files you want to process. You can drad'n'drop any node that contains recordings or sources from the database explorer to the ''Processes ''list. |
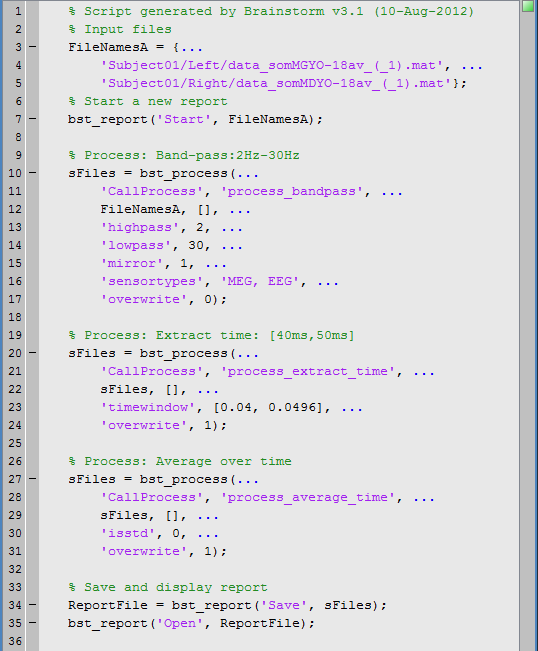
1. Drag and drop the following nodes in the Process1 list: Right/ERF (recordings), Left (condition), and Subject01 (subject)<<BR>><<BR>> {{attachment:files1.gif}} * A number between brackets appear next to each node: it represents the number of data files that where found in each of them. The node ERF "contains" only itself (1), Subject01/Right contains ERF and Std files (2), and Subject01 contains 2 conditions x 2 recordings (4). * The total number of files, ie. the sum of all those values, appears in the title of the panel "Files to process [8]". 1. The buttons on the left side allow you to select what type of file you want to process: Recordings, sources, time-frequency, other. Now select the second button "Sources". All the counts are updated and now reflect the number of sources files that are found for each node<<BR>><<BR>> {{attachment:files2.gif}} 1. If you select the third button "Time-frequency", you would see "0" everywhere because there are no time-frequency decompositions in the database yet.<<BR>><<BR>> {{attachment:files3.gif}} 1. Now clear the list from all the files. You may either right-click on the list (popup menu ''Clear list''), or select all the nodes (holding ''Shift ''or ''Crtl ''key) and then press the ''Delete ''key. 1. Select both files Left/ERF and Right/ERF in the tree (holding ''Ctrl ''key), and put the in Process list. We are going to apply some functions on those two files. You cannot distiguish them after they are dropped in the list, because they are both referred as "ERP". If at some point you need to know what is in the list, just leave you mouse over a node for a few seconds, and a tooltip would give you information about it. Just like in the database explorer.<<BR>><<BR>> {{attachment:files4.gif}} |
| Line 11: | Line 17: |
| * Click on sources file: Subject01 / !StimRightThumb / ERP / MN:MEG(Kernel). Drag'n'drop it to the empty list in ''Processes ''list. * Click on condition: Subject01 / !StimLeftThumb. Add it to the list. * Click alternatively on the ''Sources'' and ''Recordings'' buttons in the ''Processes'' tab. This indicates if you are going to process sources or recordings files. Observe that the number of files (the numbers indicated in brackets) changes according to the modality selected.<<BR>><<BR>> {{attachment:panelProcesses.gif}} * Note that you can put almost anything in this list: recordings, sources, conditions, subjects, even the whole procotol. It will take recursively all the recordings or sources files that are contained in each node. |
== Creating a pipeline == === List of processes === 1. Click on Run. The Process selection window appears, with which you can create an analysis pipeline (ie. a list of process that are applyied on the selected files one after the other). The first button in the toolbar shows the list of processed that are currently available. If you click on a menu, it's added to the list. <<BR>><<BR>> {{attachment:pipeline1.gif}} 1. Some menus appear in grey (example: Normalize > Spatial smoothing). This means that they are not meant to be applied to the type of data that you have in input, or at the end of the current pipeline. The "spatial smoothing" process may only be run on source files. 1. When you select a process, a list of options specific to this process is shown in the window. * To delete a process: Select it and press the ''Delete'' key, or the big cross in the toolbar. * With the "up arrow" and "down arrow" buttons in the toolbar, you can move up/down a process in the pipeline. 1. Now add the following processes, and set their options: * '''Pre-process > Band-pass filter''': 2Hz - 30Hz * In some processes, you can specify the type(s) of sensors on which you want to apply the process. This way you can for instance apply different filters on the EEG and the MEG, if you have both in the same files. * '''Extract > Extract time''': 40.00ms - 49.60ms, overwrite initial file * This will extract from each file a small time window around the main response peak. * Selecting the overwrite option will replace the previous file (bandpass), with the output of this process (bandpass+extract). This option is usually unselected for the first process in the list, then selected automatically. * '''Average > Average over time''': Overwrite initial file * Compute the average over this small time window. * '''Export > Export to Matlab variable''': "avg45" * Reads the current file (time window averaged over time) and exports it in a variable in the Matlab workspace.<<BR>><<BR>> {{attachment:pipeline2.gif}} 1. Save your pipeline: Click on the last button in the toolbar > Save > process_avg45. |
| Line 16: | Line 37: |
| == Process selection == * Select ''Sources''. Click on ''Run''. This big window appears:<<BR>><<BR>> {{attachment:panelStatRun.gif}} * '''Processes selection''': list of processes available for the selected data: * __Comment__: String that will be displayed in the database explorer. A " | " before means that the following string ("zscore") will be appened to the original file comment. * __Process files__: One input file => one output file * '''Z-score noise normalization''': For each input file, substract the average and divide by the variance of the baseline. * '''Remove baseline mean (DC offset)''': For each input file, substract the average of the baseline. * '''Bandpass filtering''': Hi-pass, low-pass, band-pass filtering using the Signal Processing Toolbox. * '''Spatial smoothing''': Smooth the sources activations on the cortex surface. Useful before comparing different runs or different subjects, to correct possible small errors of localization. * '''Opposite values''': Just replace the input values with their opposite * __Averaging__: Many input files => a few averaged output files * '''Average by condition''': One output for each condition (ie. grand average) * '''Average by subject''': One output for each subject * '''Average everything''': One output file only * __Extract data__: Get a structured subset of the input data (sources, scouts, clusters, ...) * '''Average over a time window''': Select a block of data in each input file, and average over the time dimension. * '''Variance over a time window''': Same, but get the variance instead of the mean. * '''Extract data block''': Select a block of data in each input file. * '''Time windows''': * __Time window__: Define the time instants that will be processed. * __Baseline__: Some processes require the definition of a baseline / pre-stimulus time window for all the input files. * '''Sources''': * Use absolute values of sources activations. If checked, the process will be applied on the absolute values of the input files, instead of the initial values.<<BR>>This is usually indispensable when processing sources amplitudes, as the relative values given by the minimum estimation are not really meaningful. * '''Scouts''': * When scouts are available in the "Scout" tab of the Brainstorm interface, they also appear here. * Check the "Use only selected scouts" if you want to compute only the scouts time series instead of extracting all the sources time series. * Check the "Merge selected scouts" if you want the selected scouts to be concatenated and considered as only one big scout. * '''Output''': * __Brainstorm database__: The results of the processes are stored in new files that will appear in the Brainstorm database explorer. <<BR>>This option is not always available, because some processes produce output that are not compatible with Brainstorm database structure. Example: If you extract the average of a scout over a certain time window, this cannot be stored in the database. * '''Overwrite initial files''': Check this option if you are applying pre-processing functions on recordings and if you don't want to keep the original files. * '''Select output condition''': A meaningful default path (Subject/Condition) to store the output files is usually selected by the program, according to the process and files processed. <<BR>>Check this option if you want to override Brainstorm's default, and enter you own output path. * __User defined file__: Store all the output information in one unique file, that has to be located ''__outside__'' of the Brainstorm database folder. * Possible output formats: Matlab .mat, or ASCII matrices * This options is usually useful if you are are extracting blocks of information that you want to process later with another software (R, Excel, Statistica), or with your own Matlab scripts. * For example: Extract 5 time windows x 3 scouts x 10 conditions x 60 subjects => You would have to start 5 times only the process and create 5 different files. * __Export to a Matlab variable__: Produce exactly the same output as the previous option, but export it directly into the current Matlab workspace instead of saving it to a file. |
=== Saving/exporting a pipeline === The last button in the the toolbar offers a list of menus to save, load and export the pipelines. |
| Line 53: | Line 40: |
| == Examples == === Z-Score noise normalization === * Keep the file selection done at the beginning of this tutorial. Select the "z-score noise normalization" process. * Set the baseline to [-49.6, -12]ms (after 12ms, the values are linear interpolations due to the electric stimulation artifact removal) * Leave the other options to their default values. Click on Run. Wait until the progress bar disappears. * Have a look to the database explorer: Two new files appeared, one for each input file.<<BR>> {{attachment:ex1_db.gif}} * For !StimRightThumb: open the ERP time series, the original sources, and the zscore.<<BR>> {{attachment:ex1_ts.gif}} {{attachment:ex1_sources.gif}} {{attachment:ex1_zscore.gif}} * Notice that the two cortical views are displayed with different colorbars. * The initial minimum norm result is shown in pA.m (amplitude range around 10e-12), with the the "sources" colormap. * The z-score values are shown without any unit, with the "stat" colormap. At a given time, a value of 20 for a given source means that its amplitude is 20 times superior to its average amplitude during the baseline. |
. {{attachment:pipeline3.gif}} |
| Line 64: | Line 42: |
| === Scout time series === * We will now extract the average value around the main response peak (41ms - 50ms), for both scouts (LeftSS, RightSS), for both conditions (StimLeftThumb, StimRightThumb). * Close all the windows * Remove the previous file selection in Processes tab (right click > Clear sample files) * Drag'n'drop the whole subject node into the Processes list (node "Subject01"), to process all the sources from Subject01 * Click on Run. There are different types of "sources" files available for Subject01, so you will be asked what files you want to process (only regular minimum norm results, zscore files, or everything). Select "Normal". * Edit the process options: * '''Process type''': Average over a time window * '''Time window''': 40.80ms - 49.60ms * '''Sources''': Keep checked the option "Use absolute values" (because we are doing a time average of sources; averaging relative values might mask some sources) * '''Scouts''': Check "Use only selected scouts", and select both scouts (LeftSS, RightSS) * '''Output''': Matlab variable * Click on Run. Enter "peak_values" as the output variable name. * In Matlab command window, type "peak_values", to see what is the content of the output structure |
* '''Load''': List of processes that are saved in the user preferences * '''Load from file''': Import a pipeline from a pipeline_...mat file (previously saved with the menu "Save as Matlab matrix") * '''Save''': Save the pipeline in the user preferencies, to be able to access it really fast after * '''Save as Matlab matrix''': Exports the pipeline for a Matlab structure in a .mat file. Allows different users to exchange their analysis pipelines (or a single user between different computers). * '''Save as Matlab script''': This option generates a human-readable Matlab script that can be re-used for other purposes or modified. * '''Delete''': Remove a pipeline that is saved in the user preferences. |
| Line 79: | Line 49: |
| {{{ >> peak_values peak_values = ImagingKernel: [] ImageGridAmp: [2x2 double] Whitener: [151x151 double] nComponents: 1 Comment: 'MN: MEG | timemean(41ms,50ms)' Function: 'minnorm' Time: [114 125] ImageGridTime: [0.0408 0.0496] ChannelFlag: [182x1 double] GoodChannel: [1x151 double] HeadModelFile: '/Subject01/StimRightThumb/headmodel_surf_os_meg_CD.mat' SurfaceFile: '/Subject01/tess_cortex_concat.mat' DataFile: '/Subject01/StimRightThumb/data_...mat' DescFileName: {'/Subject01/StimRightThumb/results_...mat' '/Subject01/StimLeftThumb/results_...mat'} DescCluster: {2x2 cell} DescProcessType: 'timemean' }}} * All the fields but the last three ones are the fields that where present in the sources files that you precessed. Among them: * '''!ImageGridAmp''': [Nscouts x Nfiles] matrix. <<BR>>The function that is used to combine the different sources into a single scout value is the function selected in the Scout tab, menu View > Time series options. Cf. previous tutorial. * '''!ImageGridTime''': Time window that was used. |
Here is the Matlab script that is generated automatically for this pipeline. |
| Line 103: | Line 51: |
| * Three fields were added to describe where the data came from, ie. the meaning of the !ImageGridAmp rows and columns.<<BR>>They all have the same number of rows and / or the same number of columns as the !ImageGrindAmp matrix. * '''!DescFileName''': [1 x Nfiles], Explains from which file came from each column of !ImageGridAmp. * '''!DescCluster''': [Nscouts x Nfiles], Explains from which cluster / scout came each value in !ImageGridAmp. * '''!DescProcessType''': Name of the process that was applied. * By looking at these fields, we can understand that the [2 x 2] matrix in !ImageGridAmp represents the average values between 40.8ms and 49.6ms for: |
. {{attachment:script.gif}} |
| Line 109: | Line 53: |
| {{{ [ StimRightThumb / LeftSS, StimLeftThumb / LeftSS; StimRightThumb / RightSS, StimLeftThumb / RightSS] }}} === Average recordings === * Let's now average all the recordings files we have for Subject01, even if it doesn't make any sense. It is just to illustrate the averaging process. * Select the files to process: * Keep the "Subject01" node in the Processes list (from previous example) * Select the "Recordings" button in the Processes tab. The "[4]" you see after Subject01 informs you that you are about to process four files (Right/ERP, Right/Std, Left/ERP, Left/Std). * Click on Run. Edit the processes options: * Process: Average by subject, or Average everything (both will lead to the same result as there is only one subject) * Keep the defaults for the other options. * Click on Run. * What happens with the database: * As you average recordings from two different conditions, the result cannot be stored in any of the existing condition. * A new condition will appear the tree, ''(Intra-subject)'', to store data that is not anymore connected to any specific condition or run, but that is still dependent only on ''Subject01''. * This ''(Intra-subject)'' node, as all the other ''"condition"'' nodes in Brainstorm database, needs a "channel file" that describes the sensors. But there is by default no channel file in this new node. * What Brainstorm offers as a very simple fix, is to create a new channel file by averaging the positions of the sensors from the conditions involved in the average. This is why you see this dialog box:<<BR>> {{attachment:combineChannel.gif}} * Click on Yes. It will average the channel files from !StimLeftThumb and !StimRightThumb conditions to create the channel file in ''(Intra-subject)''.<<BR>> {{attachment:intraSubject.gif}} * Do not try to understand the meaning of the values in ''<Subject01>'' file. We averaged recordings and variances from two different conditions: it doesn't make any sense. * '''__Warning__''': Averaging the positions of the sensors is not an accurate way to average recordings from different runs. If the position of the head within the sensors array changed a lot between the two runs, this method can introduce an important error, because you average fields that do not come from the same position in space. * If you really want to average MEG recordings coming from different runs properly, you have to register them with dedicated tools, that transform the fields recorded and make them spatially compatible. * Brainstorm will be able to perform this operation soon, but for the moment you still have to do that with another program. * This "channel file average" is just a quick and inaccurate tool to allow the users to compute an average in case the head of the subject did not move to much between the runs. * To check the difference between sensors positions in two runs: select all the channel files at the same time, right click, menu Display sensors.<<BR>> {{attachment:comparePositions.gif}} {{attachment:comparePositionsChan.gif}} |
=== Running a pipeline === 1. Click on Ok, in the pipeline window. After a few seconds, you will see two new files in the database, and two messages in the Matlab command window. The file names have list of tags that where appended to represent all the processes that were applied to them. <<BR>><<BR>> {{attachment:output.gif}} {{attachment:export.gif}} 1. Quick reminder: the file history record all this information too (right-click > File > View file history), so you are free to rename those files in something shorter (example: "ERF_avg45").<<BR>><<BR>> {{attachment:history.gif}} 1. Now double-click on the processed file in Right condition. * You see only horizontal lines. It is not a bug, it is because you averaged the data in time. There is no temporal dimension anymore. * Press Ctrl+T, to bring up a "2D sensor cap" view. It represents the average topography between 40ms and 50ms, filtered between 2Hz and 30Hz.<<BR>><<BR>> {{attachment:fig_avg.gif}} 1. Now have a look to the output in the command window. Two variables where created, because there were two files in output "avg45_1" and "avg_45_2". Type "avg45_1" in the command window to see the fields in the structure:<<BR>><<BR>> {{attachment:avg45.gif}} * This is the exact same structure as the processed file in condition Left. All those fields where documented in the tutorial #2. Just notice the dimension of the recordings matrix F: [182 x 2], and the Time vector: [0.0400 0.0496] * In a general way, all the averages in time are represented in Brainstorm by two identical columns, one for the beginning and one for the end of the time window they represent. 1. Delete the two processed files, they are of no use after. == Another example: z-score and scouts == 1. Load the two scouts LeftSS and RightSS saved in the previous tutorial. 1. Clear the files list and drag'n'drop "Subject01". 1. Select the "Results" button of the Process1 panel, it should indicate two files. Click on run. 1. Create the following pipeline: * Normalize > Z-score noise normalization: baseline = [-50ms,-1ms] * Extract > Scouts time series: select both scouts, and select "Concatenate output in one matrix"<<BR>><<BR>> {{attachment:pipeline_zscore.gif}} 1. Run the pipeline. It creates three new files in the database: 1. {{attachment:zscore_output.gif}} Let's have a detailed look at each of those files: * '''Left / ERF / MN | zscore''': Output of the zscore process for the Left sources * Double-click on it to display it. Display the original sources at the same time<<BR>><<BR>> {{attachment:zscore_cortex.gif}} * The z-score operation substracts the average and divide by the variance of the baseline. The goal is to give less importance to the sources that havea strong variability in the baseline, because they are considered as noise, and this way increase the visibility of the significant effects. * The maps look relatively similar in terms of localization, except that for the z-score, we do not observe any pattern of activity in the poles of the temporal lobes. The variance in those areas was high during the baseline, they have been "removed" by the z-score process. * Note that the units changed. The values of the z-score are without units (the minimum norm values where divided by their standard deviation) * '''Right / ERF / MN | zscore''': Output of the zscore process for the Right sources. Same thing. * '''Extract scouts: LeftSS RightSS [2 files]''': Scouts times series for the two zscore files * This file was saved in a new node in the database: "Intra-subject". This is where are stored the results of processes that have in inputs files from different conditions within the same subject. * It contains the concatenated output of 2 scouts x 2 conditions, in one unique matrix. If we had not selected the "Concatenate" option, two files would have been saved, one in condition Left, one in condition Right. * This is a type of file that was not introduced yet in those tutorials. It is called simply "matrix", and may contain any type of data. Because we don't exactly know what they contain, the operations that can be performed on them are very limited. . {{attachment:matrix_popup.gif}} {{attachment:matrix_view.gif}} Last thing to do before going to the next tutorial: delete those three files. |
| Line 136: | Line 93: |
| Comparisons between different conditions with the [[Tutorials/TutStat|Statistics]] tab. |
[[Tutorials/TutStat|Process with two sets of input]]. |
Tutorial 9: Processes
The main window includes a graphical batching interface that directly benefits from the database explorer: files are organized as a tree of subjects and conditions, and simple drag-and-drop operations readily select files for subsequent batch processing. Most of the Brainstorm features are (or will be) available through this interface, including pre-processing of the recordings, averaging, time-frequency decompositions, and computing statistics. A full analysis pipeline can be created in a few minutes, saved in the user’s preferences and reloaded in one click, executed directly or exported as a Matlab script.
The available processes are organized in a plug-in structure. Any Matlab script that is added to the plug-in folder (brainstorm3/toolbox/process/functions/) and has the right format will be automatically detected and made available in the GUI. This mechanism makes the contribution from other developers to Brainstorm very easy.
Selecting files to process
First thing to do is to define the files you are going to process. This is done easily by picking files or folders in the database explorer and droping them in the empty list of the Process1 tab.
Drag and drop the following nodes in the Process1 list: Right/ERF (recordings), Left (condition), and Subject01 (subject)
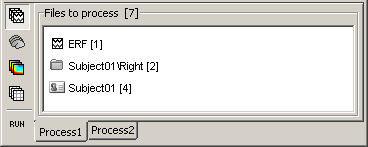
- A number between brackets appear next to each node: it represents the number of data files that where found in each of them. The node ERF "contains" only itself (1), Subject01/Right contains ERF and Std files (2), and Subject01 contains 2 conditions x 2 recordings (4).
- The total number of files, ie. the sum of all those values, appears in the title of the panel "Files to process [8]".
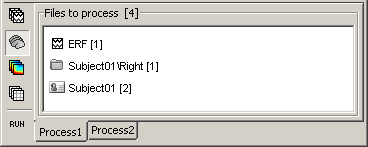
The buttons on the left side allow you to select what type of file you want to process: Recordings, sources, time-frequency, other. Now select the second button "Sources". All the counts are updated and now reflect the number of sources files that are found for each node
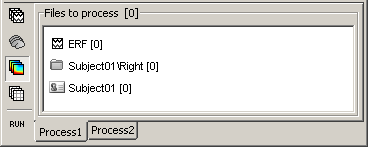
If you select the third button "Time-frequency", you would see "0" everywhere because there are no time-frequency decompositions in the database yet.
Now clear the list from all the files. You may either right-click on the list (popup menu Clear list), or select all the nodes (holding Shift or Crtl key) and then press the Delete key.
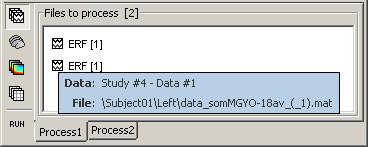
Select both files Left/ERF and Right/ERF in the tree (holding Ctrl key), and put the in Process list. We are going to apply some functions on those two files. You cannot distiguish them after they are dropped in the list, because they are both referred as "ERP". If at some point you need to know what is in the list, just leave you mouse over a node for a few seconds, and a tooltip would give you information about it. Just like in the database explorer.
Creating a pipeline
List of processes
Click on Run. The Process selection window appears, with which you can create an analysis pipeline (ie. a list of process that are applyied on the selected files one after the other). The first button in the toolbar shows the list of processed that are currently available. If you click on a menu, it's added to the list.
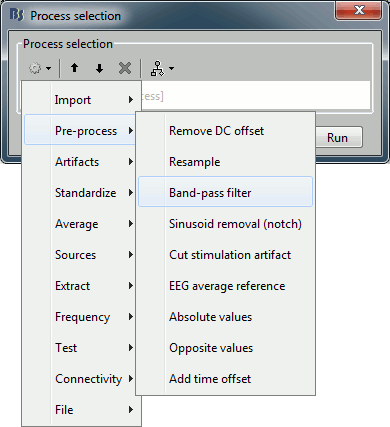
Some menus appear in grey (example: Normalize > Spatial smoothing). This means that they are not meant to be applied to the type of data that you have in input, or at the end of the current pipeline. The "spatial smoothing" process may only be run on source files.
- When you select a process, a list of options specific to this process is shown in the window.
To delete a process: Select it and press the Delete key, or the big cross in the toolbar.
- With the "up arrow" and "down arrow" buttons in the toolbar, you can move up/down a process in the pipeline.
- Now add the following processes, and set their options:
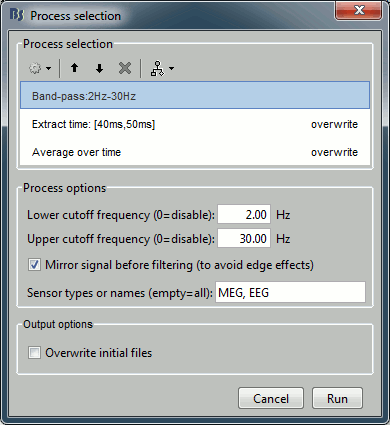
Pre-process > Band-pass filter: 2Hz - 30Hz
- In some processes, you can specify the type(s) of sensors on which you want to apply the process. This way you can for instance apply different filters on the EEG and the MEG, if you have both in the same files.
Extract > Extract time: 40.00ms - 49.60ms, overwrite initial file
- This will extract from each file a small time window around the main response peak.
- Selecting the overwrite option will replace the previous file (bandpass), with the output of this process (bandpass+extract). This option is usually unselected for the first process in the list, then selected automatically.
Average > Average over time: Overwrite initial file
- Compute the average over this small time window.
Export > Export to Matlab variable: "avg45"
Reads the current file (time window averaged over time) and exports it in a variable in the Matlab workspace.
Save your pipeline: Click on the last button in the toolbar > Save > process_avg45.
Saving/exporting a pipeline
The last button in the the toolbar offers a list of menus to save, load and export the pipelines.
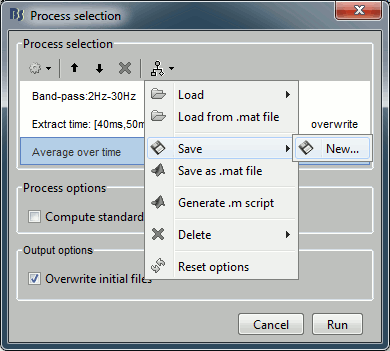
Load: List of processes that are saved in the user preferences
Load from file: Import a pipeline from a pipeline_...mat file (previously saved with the menu "Save as Matlab matrix")
Save: Save the pipeline in the user preferencies, to be able to access it really fast after
Save as Matlab matrix: Exports the pipeline for a Matlab structure in a .mat file. Allows different users to exchange their analysis pipelines (or a single user between different computers).
Save as Matlab script: This option generates a human-readable Matlab script that can be re-used for other purposes or modified.
Delete: Remove a pipeline that is saved in the user preferences.
Here is the Matlab script that is generated automatically for this pipeline.
Running a pipeline
Click on Ok, in the pipeline window. After a few seconds, you will see two new files in the database, and two messages in the Matlab command window. The file names have list of tags that where appended to represent all the processes that were applied to them.
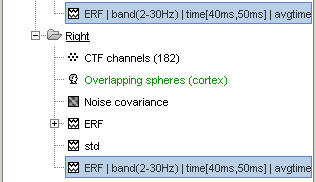

Quick reminder: the file history record all this information too (right-click > File > View file history), so you are free to rename those files in something shorter (example: "ERF_avg45").
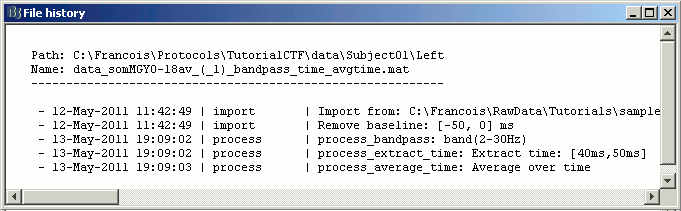
- Now double-click on the processed file in Right condition.
- You see only horizontal lines. It is not a bug, it is because you averaged the data in time. There is no temporal dimension anymore.
Press Ctrl+T, to bring up a "2D sensor cap" view. It represents the average topography between 40ms and 50ms, filtered between 2Hz and 30Hz.
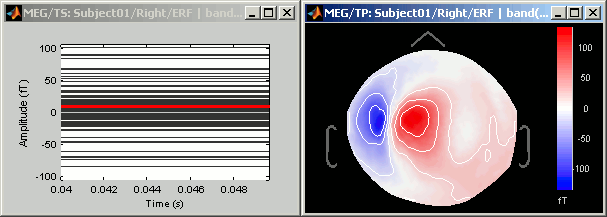
Now have a look to the output in the command window. Two variables where created, because there were two files in output "avg45_1" and "avg_45_2". Type "avg45_1" in the command window to see the fields in the structure:
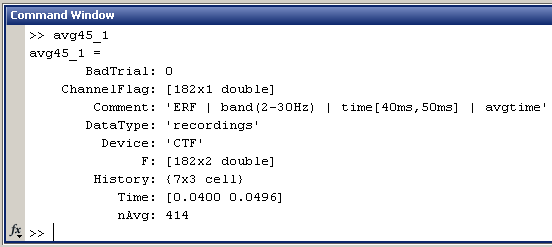
- This is the exact same structure as the processed file in condition Left. All those fields where documented in the tutorial #2. Just notice the dimension of the recordings matrix F: [182 x 2], and the Time vector: [0.0400 0.0496]
- In a general way, all the averages in time are represented in Brainstorm by two identical columns, one for the beginning and one for the end of the time window they represent.
- Delete the two processed files, they are of no use after.
Another example: z-score and scouts
- Load the two scouts LeftSS and RightSS saved in the previous tutorial.
- Clear the files list and drag'n'drop "Subject01".
- Select the "Results" button of the Process1 panel, it should indicate two files. Click on run.
- Create the following pipeline:
Normalize > Z-score noise normalization: baseline = [-50ms,-1ms]
Extract > Scouts time series: select both scouts, and select "Concatenate output in one matrix"
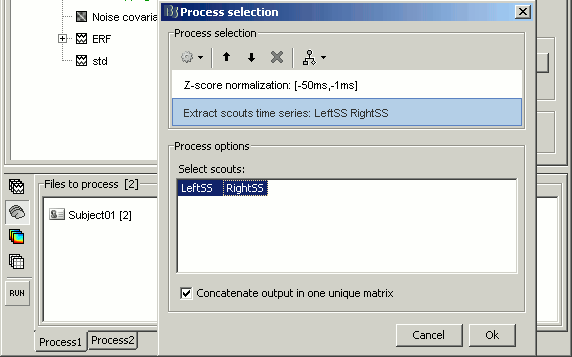
- Run the pipeline. It creates three new files in the database:
Let's have a detailed look at each of those files:
Left / ERF / MN | zscore: Output of the zscore process for the Left sources
Double-click on it to display it. Display the original sources at the same time
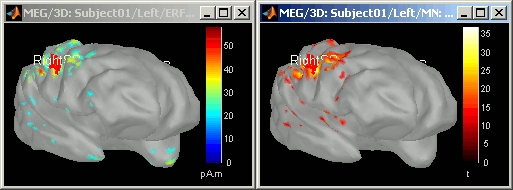
- The z-score operation substracts the average and divide by the variance of the baseline. The goal is to give less importance to the sources that havea strong variability in the baseline, because they are considered as noise, and this way increase the visibility of the significant effects.
- The maps look relatively similar in terms of localization, except that for the z-score, we do not observe any pattern of activity in the poles of the temporal lobes. The variance in those areas was high during the baseline, they have been "removed" by the z-score process.
- Note that the units changed. The values of the z-score are without units (the minimum norm values where divided by their standard deviation)
Right / ERF / MN | zscore: Output of the zscore process for the Right sources. Same thing.
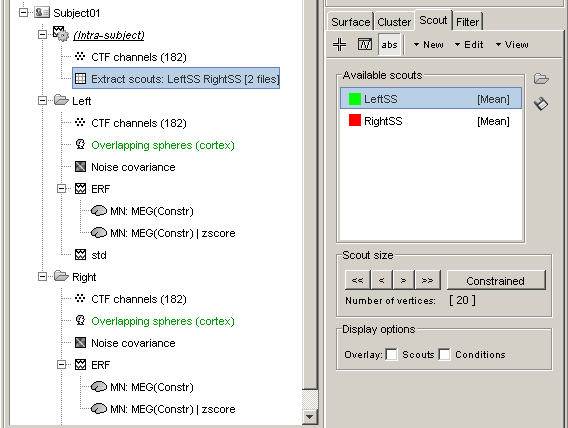
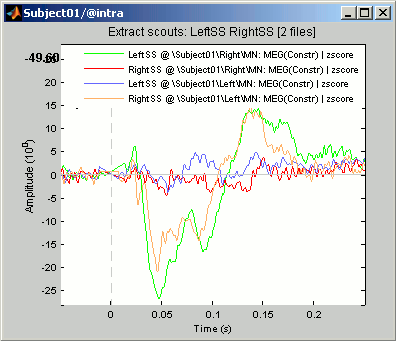
Extract scouts: LeftSS RightSS [2 files]: Scouts times series for the two zscore files
- This file was saved in a new node in the database: "Intra-subject". This is where are stored the results of processes that have in inputs files from different conditions within the same subject.
- It contains the concatenated output of 2 scouts x 2 conditions, in one unique matrix. If we had not selected the "Concatenate" option, two files would have been saved, one in condition Left, one in condition Right.
- This is a type of file that was not introduced yet in those tutorials. It is called simply "matrix", and may contain any type of data. Because we don't exactly know what they contain, the operations that can be performed on them are very limited.
Last thing to do before going to the next tutorial: delete those three files.
Next
?Process with two sets of input.