|
Size: 3336
Comment:
|
Size: 4291
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 7: | Line 7: |
| The raw file viewer provides a rapid access to the recordings, but most of operations cannot be performed directly on the continuous files: most of the pre-processing functions, averaging, time-frequency analysis and statistical tests can only be applied on blocks of data that are saved in the database (ie. "imported"). After reviewing the recordings and editing the event markers, you have to "import" the recordings in the database to go with further analysis. The two following operations produce the same result: | The raw file viewer provides a rapid access to the recordings, but most of operations cannot be performed directly on the continuous files: most of the pre-processing functions, averaging, time-frequency analysis and statistical tests can only be applied on blocks of data that are saved in the database (ie. "imported"). After reviewing the recordings and editing the event markers, you have to "import" the recordings in the database to go with further analysis. |
| Line 9: | Line 9: |
| * In the Event tab: select the menu Import > Import > Import in database (only when the file viewer is open), * In the database tree: Right-click on the nodes "Link to raw file" > Import in database. If you right-click on the subject node > Import EEG/MEG, and select the tutorial .ds dataset, you would be able to import blocks of the continuous file in the database, but you would not have access to the modified events list or the SSP operators. Therefore, you would not import data cleaned for the ocular and cardiac artifacts. The modified events list and the signal space projectors are saved only in the "Link to raw file" in the Brainstorm database, not in the initial continuous file. Now, right-click on the file with power line correction:''' Raw | sin(60Hz 120Hz 180Hz) > Import in dabase''' |
Right-click on the file with power line correction:''' Raw | sin(60Hz 120Hz 180Hz) > Import in dabase''' |
| Line 17: | Line 12: |
__Warning__: If you right-click on the subject node > Import EEG/MEG, and select the tutorial .ds dataset, you would be able to import blocks of the continuous file in the database, but you would not have access to the modified events list or the SSP operators. Therefore, you would not import data cleaned of ocular and cardiac artifacts. The modified events list and the signal space projectors are saved only in the "Link to raw file" in the Brainstorm database, not in the initial continuous file. |
|
| Line 19: | Line 17: |
| * '''Time window''': Time range of interest. Now we want to keep the whole time definition, we are interested by all the stimulations. * '''Split''': Useful to import continuous recordings without events. We do not need this here. * '''Events selection''': Check the "Use events" option, and select both STI001 and STI002. The value between parenthesis represents the number of occurrences of this event. * '''Epoch time''': Time instants that will be extracted before an after each event, to create the epochs that will be saved in the database. Set it to [-100, +300] ms * '''Use Signal Space Projections''': Save and use the SSP vectors created by MaxFilter or during other pre-processing steps. Keep this option selected. |
Set the import options as they are represented in this figure: * '''Time window''': Time range of interest. Now we want to keep the whole time definition, we are interested by all the stimulations, so leave the default values * '''Split''': Useful to import continuous recordings without events, to import successive chunks of the same duration. We do not need this here. * '''Events selection''': Check the "Use events" option, and select both "left" and "right". The value between parenthesis represents the number of occurrences of this event in the selected time window (would change if you modify the time definition on top of the figure) * '''Epoch time''': Time segment that is extracted around each marker, to create the epochs that are saved in the database. Set it to [-100, +300] ms * '''Use Signal Space Projections''': Use the active SSP projectors calculated during the previous pre-processing steps. Keep this option selected. |
| Line 26: | Line 26: |
| * '''Create new conditions for epochs''': If selected, a new condition is created for each event type (here, it will create a condition "Event_5"). If not selected, all the epochs from all the selected events are saved in the same condition, the one that was selected in the database explorer (if no condition selected, create a condition called "New condition"). * Click on Import and wait. |
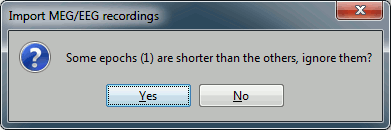
* '''Create new conditions for epochs''': If selected, a new condition is created for each event type (here, it will create two conditions in the database: "left" and "right"). If not selected, all the epochs are saved in a new folder, the same one for all the events, that has the same name as the initial raw file. Click on Import and wait. In the end, you are asked whether you want to ignore one epoch that is shorter than the others. This happens because the acquisition of the MEG signals was stopped less than 300ms after the last stimulus trigger was sent. Therefore, the last epoch cannot have the full [-100,300]ms time definition. This shorter epoch would prevent us from averaging all the epochs easily. As we already have enough repetitions in this experiment, we can ignore it. Answer '''Yes''' to this question to discard the last epoch. |
| Line 31: | Line 32: |
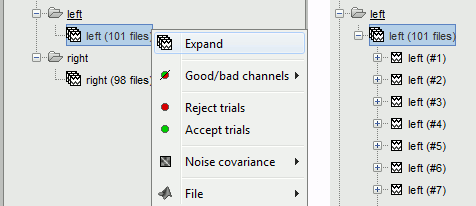
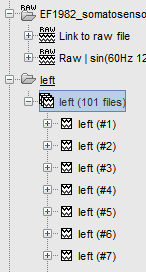
| Two new conditions containing two groups of trials appeared in the database. The groups of trials are not expanded by default, to have the database tree displayed faster. To expand a list and get access to the individual trials: double click on it, or right-click on the list > Expand. | |
| Line 32: | Line 34: |
| {{attachment:importDb.gif}} | The SSP projectors calculated in the previous tutorial were applied on the fly when reading from the continuous file. Those epochs are clean from eye blinks and power line contamination. {{attachment:importDb.gif}} {{attachment:importDbExpand.gif|importDb.gif}} |
Epoching and averaging
This tutorial fills the gap between the previous tutorial (review and clean continuous recordings), and the introduction tutorials (source analysis of evoked responses). It explains how to epoch the recordings, do some more pre-processing on the single trials, and then calculate their average.
Contents
Import in database
The raw file viewer provides a rapid access to the recordings, but most of operations cannot be performed directly on the continuous files: most of the pre-processing functions, averaging, time-frequency analysis and statistical tests can only be applied on blocks of data that are saved in the database (ie. "imported"). After reviewing the recordings and editing the event markers, you have to "import" the recordings in the database to go with further analysis.
Right-click on the file with power line correction: Raw | sin(60Hz 120Hz 180Hz) > Import in dabase
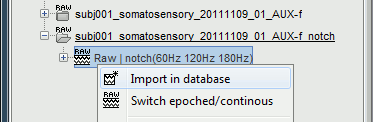
Warning: If you right-click on the subject node > Import EEG/MEG, and select the tutorial .ds dataset, you would be able to import blocks of the continuous file in the database, but you would not have access to the modified events list or the SSP operators. Therefore, you would not import data cleaned of ocular and cardiac artifacts. The modified events list and the signal space projectors are saved only in the "Link to raw file" in the Brainstorm database, not in the initial continuous file.
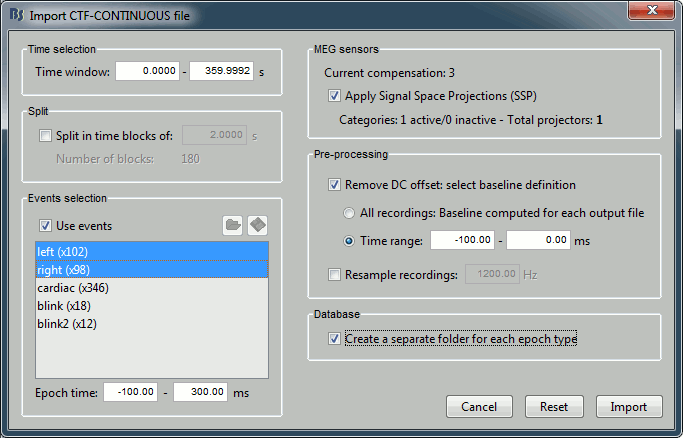
Set the import options as they are represented in this figure:
Time window: Time range of interest. Now we want to keep the whole time definition, we are interested by all the stimulations, so leave the default values
Split: Useful to import continuous recordings without events, to import successive chunks of the same duration. We do not need this here.
Events selection: Check the "Use events" option, and select both "left" and "right". The value between parenthesis represents the number of occurrences of this event in the selected time window (would change if you modify the time definition on top of the figure)
Epoch time: Time segment that is extracted around each marker, to create the epochs that are saved in the database. Set it to [-100, +300] ms
Use Signal Space Projections: Use the active SSP projectors calculated during the previous pre-processing steps. Keep this option selected.
Remove DC Offset: Check this option, and select: Time range: [-100, 0] ms. For each epoch, this will: compute the average of each channel over the baseline (pre-stimulus interval: -100ms to 0ms), and subtract it from the channel at all the times in [-100,+300]ms.
Resample recordings: Keep this unchecked
Create new conditions for epochs: If selected, a new condition is created for each event type (here, it will create two conditions in the database: "left" and "right"). If not selected, all the epochs are saved in a new folder, the same one for all the events, that has the same name as the initial raw file.
Click on Import and wait. In the end, you are asked whether you want to ignore one epoch that is shorter than the others. This happens because the acquisition of the MEG signals was stopped less than 300ms after the last stimulus trigger was sent. Therefore, the last epoch cannot have the full [-100,300]ms time definition. This shorter epoch would prevent us from averaging all the epochs easily. As we already have enough repetitions in this experiment, we can ignore it. Answer Yes to this question to discard the last epoch.
Two new conditions containing two groups of trials appeared in the database. The groups of trials are not expanded by default, to have the database tree displayed faster. To expand a list and get access to the individual trials: double click on it, or right-click on the list > Expand.
The SSP projectors calculated in the previous tutorial were applied on the fly when reading from the continuous file. Those epochs are clean from eye blinks and power line contamination.